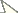(C) 2011 Rosiley Berton Pacheco. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Two populations of Astyanax altiparanae (Garutti & Britski, 2000) of the Água dos Patos stream/SP and lake Igapó/PR were analyzed. All individuals showed 2n = 50, however, different karyotypic formulae were observed. The population of the Água dos Patos stream showed 8m +24sm+6st+12a (NF=88) and the population of lake Igapó, 8m+28sm+4st+10a (NF=90). Nucleolus organizing regions (AgNORs) were observed in the terminal position on the short and long arm of different chromosomes of both populations, showing a variation from 3 to 4 chromosomes. Fluorescent in situ hybridization (FISH) using 18S rDNA probes revealed only one pair of chromosomes with fluorescent signals in the terminal site on the short arm in the Igapó lake population, while the population of Água dos Patos stream showed 4 fluorescence terminal signals, characterizing a system of simple and multiple NORs, respectively. 5S rDNA fluorescent signals were detected in the interstitial position of a pair of chromosomes in the two studied populations. Some AgNOR sites revealed to be GC-rich when stained with Chromomycin A3 (CMA3), however, AT positive regions were not observed. The data obtained show that, despite the conservation of the diploid number and location of 5S DNAr, differences in both the distribution of 18S rDNA and karyotypic formula among the populations were found, thus corroborating the existing data on chromosome variability in Astyanax altiparanae that can be significant for cytotaxonomy in this group.
Teleostei Characidae, 18S rDNA, 5S rDNA, FISH, karyotypic formula, NORs
Astyanax Baird & Girard, 1854, the most common and diversified genus within the family Characidae, has a wide distribution in the Neotropical Region. Due to lack of evidence of monophyly, the genus Astyanax is thought to belong to the Incertae sedis group (
In Astyanax altiparanae (Garutti & Britski, 2000) from the upper Parana river basin, previously identified as Astyanax bimaculatus
(Linnaeus, 1758), all cytogenetic studies accomplished so far
reported the occurrence of 2n = 50, with differences in the karyotypic
formula among the analyzed populations (
Besides the differences in karyotypic formula, the
nucleolus organizer regions in this species also vary in relation to
number and position, as observed by
In view of the great chromosome variation observed by other authors in the genus Astyanax, the objective of the present work was to characterize the karyotypes of two populations of Astyanax altiparanae, with emphasis on the location of 18S and 5S DNAr sites, and compare them with data contained in the literature, in an endeavor toward a better understanding of chromosome evolution within this fish group.
Material and methodsTwo populations of Astyanax altiparanae were cytogenetically analyzed: twelve specimens (3 males and 9 females) from Água dos Patos stream (22°41'17.7"S; 51° 05'23.9"W ), municipality of Iêpe/SP and sixteen specimens (9 males and 7 females) from Igapó lake (23°19'09.38"S; 51°11'44.72"W ), municipality of Londrina/PR (Fig. 1). Specimens were deposited in the Museum of Zoology of the Universidade Estadual de Londrina (MZUEL). The samples were collected with the permission of Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (IBAMA), protocol number 11399-1. This study was approved by the ethics committee of our institution and meets all requirements of the Brazilian environmental laws.
Collection sites of studied specimens. Map of Brazil showing the Paraná and São Paulo states in the selected area (left side). Hydrographic map showing the Paranapanema and Tibagi rivers. In (a) Água dos Patos stream and (b) Igapó lake.
Conventional staining. The specimens were
sacrificed after being anesthetized with a solution of benzocaine.
Metaphase chromosomes were obtained from kidney cells according to the
air drying technique (
Fluorescent in situ hybridization (FISH). The in situ hybridization procedure was performed according to
Chromosome banding. Active nucleolus organizer regions (NORs) were detected by silver nitrate staining (
All the images were acquired with a Leica DM 4500 B microscope equipped with a DFC 300FX camera and Leica IM50 4.0 software, and optimized for best constrast and brightness with iGrafx Image software.
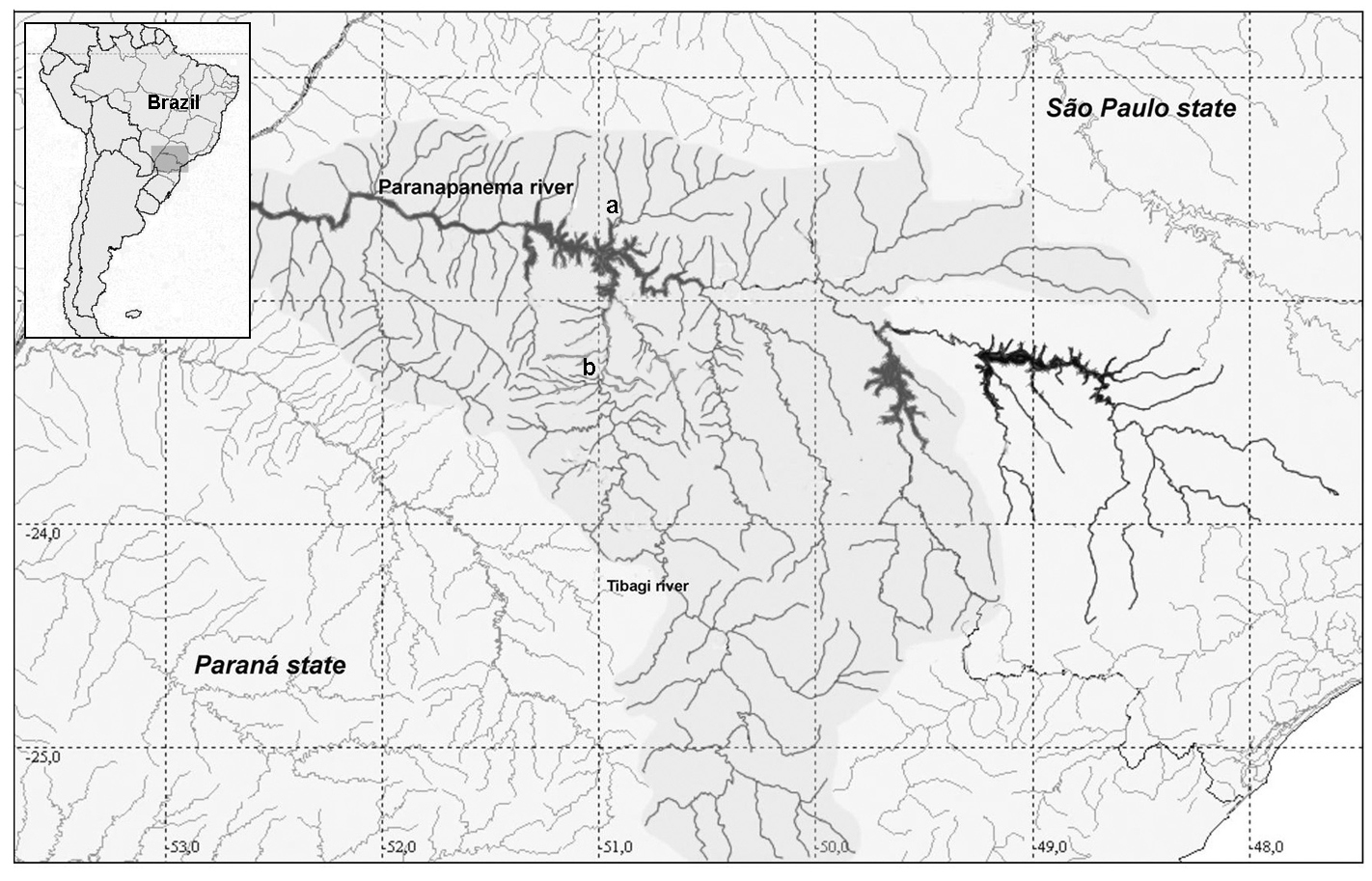
ResultsThe two populations of Astyanax altiparanae showed 2n = 50, however, different karyotypic formula were evidenced. The population of the Água dos Patos stream showed 8m+24sm+6st+12a (NF=88) (Fig. 2a) and the population of Igapó lake 8m+ 28sm+ 4st+10a (NF=90) (Fig. 2b).
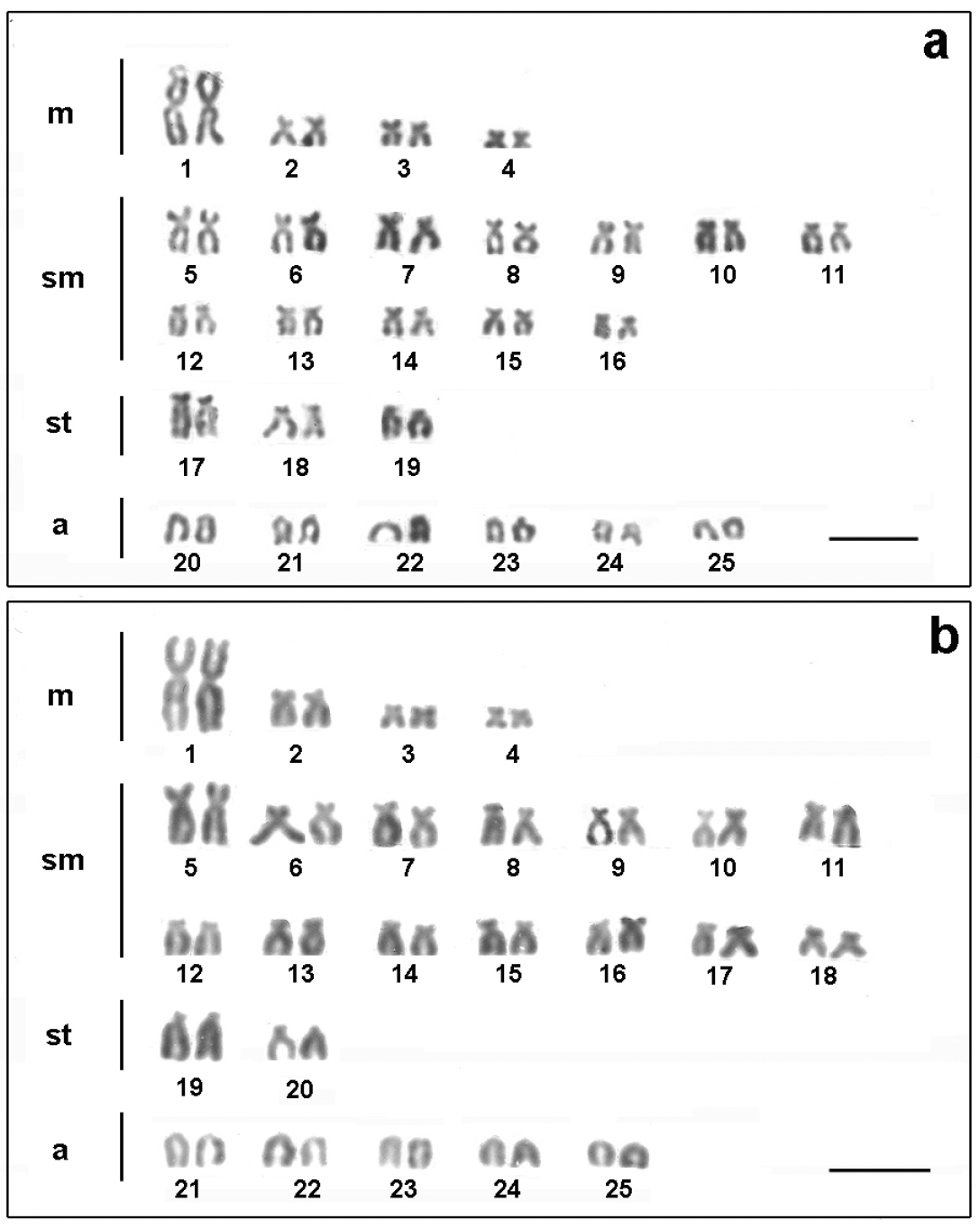
Multiple NORs sites were detected in the two populations by silver nitrate impregnation, revealing inter- and intra-individual variations. The population of the Água dos Patos stream showes 2 to 4 AgNORs on the short arm, in two equal medium-sized subtelocentric chromosomes, one of which revealed size heteromorphism observed in all metaphases (Fig. 3a).
Karyotypes of Astyanax altiparanae after conventional Giemsa staining a Água dos Patos stream b Igapó lake. Bar= 5μm.
Chromosomes of Astyanax altiparanae bearing AgNORs sites: a Água dos Patos stream e Igapó lake. Metaphase chromosomes with the arrows showing the 18S rDNA, CMA3/DAPI and 5S rDNA sites in Astyanax altiparanae, respectively: b, c, d Água dos Patos stream f, g, h Igapó lake. Bar = 5μm.
One to three AgNORs were detected in the population of Igapó lake: a medium-sized submetacentric chromosome with markings on the long arm and also a medium-sized subtelocentric chromosome with signal on the short arm, presenting a small size heteromorphism. The latter was observed in almost all metaphases (Fig. 3e).
The population of the Água dos Patos stream showed 4 markings on the short arm of two medium-sized subtelocentric chromosome pairs after fluorescent in situ hybridization (FISH) with 18S rDNA probe (Fig. 3b). In the population of Igapó lake, only one pair of medium-sized subtelocentric chromosome presenting fluorescent signals on the short arm was detected (Fig. 3f). Both populations exhibited one pair of chromosomes, bearers of the 5S DNAr sites, in the interstitial position (Fig. 3d and h).
In the population of the Água dos Patos stream, 2 to 4 chromosomes with CMA3+ terminal blocks were detected: a medium-sized subtelocentric pair with signals of size heteromorphism on the short arm, most frequently visualized in the metaphases; a small-sized acrocentric chromosome with a marking on the short arm; and a medium-sized subtelocentric chromosome with a signal on the long arm (Fig. 3c). In the population of Igapó lake, CMA3 markings were detected on the short arm of only one pair of medium-sized subtelocentric chromosomes, which disclosed size heteromorphism (Fig. 3g).
The treatment of the chromosomal preparations of the two populations with DAPI showed a homogeneous staining region, and no regions rich in AT base pairs were detected, as can be seen through the superposition of these fluorochrome, as shown in Fig. 3c-g.
DiscussionCytogenetic studies in Astyanax altiparanae from the Água dos Patos stream and Igapó lake revealed a conserved diploid number that has been observed within all the analyzed populations of this species (Table 1) so far. However, differences in the karyotypic formula were found in some of these populations, including the one observed in the present study, probably due to occurrence of chromosomal rearrangements, such as pericentric inversions, thus revealing a variability in the karyotypic macrostructure among the species of this group of fish.
Cytogenetic data of different populations of Astyanax altiparanae. FN: fundamental number; m: metacentric; sm: submetacentric; st: subtelocentric; a: acrocentric; AgNORs: nucleolar organizer regions; Ref: Reference; PR: Paraná; SP: São Paulo
| Locality | 2n | FN | Chromosome formulae | AgNORs | CMA3 | 18S/28S | 5S | Ref. |
|---|---|---|---|---|---|---|---|---|
| Mogi Guaçu river/SP | 50 | 88 | 10m+24sm+4st+12a | 1 | ||||
| 50 | 92 | 6m+24sm+12st+8a | 1 a 5 | 2 | ||||
| Paranapanema river/SP | 50 | 88 | 10m+22sm+6st+12a | 3 | ||||
| Tibagi river/Sertanópolis/PR | 50 | 90 | 10m+22sm+8st+10a | 2 a 5 | 5 | 4 | ||
| Tibagi river/Limoeiro/PR | 50 | 86 | 6m+22sm+8st+14a | 2 a 5 | 5 | 4 | ||
| Tibagi river/Limoeiro/PR | 50 | 88 | 10m+22sm+6st+12a | 2 a 5 | 5 | 4 | ||
| Couro de Boi river/PR | 50 | 88 | 8m+20sm+10st+12a | 1 a 4 | 6 | 4 | ||
| Três Bocas stream/PR | 50 | 92 | 10m+28sm+4st+8a | 1 a 6 | 8 | 4 | ||
| Claro river/PR | 50 | 90 | 10m+26sm+4st+10a | 1 a 4 | 6 | 5 | ||
| 88 | 10m+24sm+4st+12a | 1 a 4 | 6 | 5 | ||||
| 86 | 10m+22sm+4st+14a | 1 a 4 | 6 | 5 | ||||
| Paraná river/PR | 50 | 82 | 32m/sm+18st/a | 3 | 4 | 2 | 6 | |
| Índios river/PR | 50 | 90 | 6m+30sm+4st+10a | 10 | 7 | 7 | ||
| Paraná river/PR | 50 | 88 | 6m+26sm+6st+12a | 2 | 5 | 4 | 2 | 7, 9 |
| Tibagi river upper/PR | 50 | 92 | 6m+28sm+8st+8a | 4 | 7 | 7 | 2 | 8 |
| Iguaçu river upper/PR | 50 | 94 | 6m+30sm+8st+6a | 2 | 4 | 2 | 2 | 8 |
| Keçaba brook/PR | 50 | 88 | 6m+26sm+6st+12a | 3 | 7 | 2 | 9 | |
| Tatupeba brook/PR | 50 | 88 | 6m+26sm+6st+12a | 3 | 4 | 2 | 9 | |
| Maringá stream/PR | 50 | 88 | 6m+26sm+6st+12a | 1 | 4 | 2 | 9 | |
| 50 | 88 | 10m+22sm+6st+12a | 2 a 3 | 10 | ||||
| Iguaçu river/PR | 50 | 92 | 10m+26sm+6st+8a | 2 a 5 | 10 | |||
| Monjolinho river/SP | 50 | 90 | 8m+20sm+12st+10a | 2 | 2 | 2 | 11 | |
| Água dos Patos stream/SP | 50 | 88 | 8m+24sm+6st+12a | 2 a 4 | 2 a 4 | 4 | 2 | 12 |
| Igapó lake/PR | 50 | 90 | 8m+28sm+4st+10a | 1 a 3 | 2 | 2 | 2 | 12 |
References: 1
The great variability in the karyotypic macrostructure is also reflected in other chromosome marks of Astyanax altiparanae.
Variability in AgNOR sites with respect to the number, location and
types of chromosomes bearers of such sites is frequently evidenced in
this species (Table 1),
as corroborated by the present study. Some authors consider that such
variations can be ascribed to chromosomal rearrangements and transfer of
ribosomic sites (
It is worth noting that an AgNORs pair was the most
frequently found in the chromosome preparations of the populations
analyzed herein. It can be considered a main pair with NOR always
active, together with secondary sites, as observed by
After FISH with 18S rDNA probe, Astyanax altiparanae
of Água dos Patos stream showed two chromosome pairs with fluorescent
signals on the short arms, coinciding with the sites detected by silver
impregnation. The population of Igapó lake, however, showed only one
chromosome pair with fluorescent signals on the short arm, coinciding
with a pair frequently identified by silver nitrate, thus
characterizing a system of simple NORs. The other markings, which had
not been identified by FISH, but were observed in this population after
the impregnation with silver nitrate, are probably heterochromatic
sites with acid proteins that have affinity to silver. Despite the fact
that multiple NORs are a common condition among Astyanax altiparanae (Table 1),
Multiple 18S rDNA sites were also found among other species of the genus Astyanax, such as Astyanax scabripinnis (Jenyns, 1842) (
The CMA3+ sites of Astyanax altiparanae
of Igapó lake and Água dos Patos stream were consistent with those
marked by silver, however, the AgNORs not detected through CMA3 in the
individuals of Igapó lake may be very small and not detectable by this
fluorochrome, or else not all NORs are rich in GC, as suggested by
The Ag-NOR heteromorphism observed in the two populations was not seen by FISH, thence, it should be related to the expression of the genes or be ascribable to a larger amount of heterochromatin that insert in 18S DNAr cistrons, once it was detected by CMA3 and by the impregnation with silver nitrate.
After staining with DAPI, markings were not observed in the chromosomes of the two populations of Astyanax altiparanae, which, therefore, did not possess any region rich in AT bases.
In all the populations of Astyanax altiparanae,
the5S DNAr sites were located interstitially in one chromosome pair,
demonstrating a high stability of those sites. This corroborates the
data on other populations of Astyanax altiparanae (Table 1) and of other species of the genus Astyanax (
According to
The authors are grateful to CAPES, CNPq and Fundação Araucária for financial support and to Dr. Oscar A. Shibatta for the identification of the studied species.