(C) 2012 Nomar Espinosa Waminal. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Ginseng has long been considered a valuable plant owing to its medicinal properties; however, genomic information based on chromosome characterization and physical mapping of cytogenetic markers has been very limited. Dual-color FISH karyotype and DAPI banding analyses of Panax ginseng C. A. Meyer, 1843 were conducted using 5S and 45S rDNA probes. The somatic chromosome complement was 2n=48 with lengths from 3.3 μm to 6.3 μm. The karyotype was composed of 12 metacentric, 9 submetacentric, and 3 subtelocentric pairs. The 5S rDNA probe localized to the intercalary region of the short arm of pair 11, while the 45S rDNA was located at the secondary constriction of the subtelocentric satellited chromosome 14. DAPI bands were clearly observed for most chromosomes, with various signal intensities and chromosomal distributions that consequently improved chromosome identification. As a result, all 24 chromosomes could be distinguished and numbers were assigned to each chromosome for the first time. The results presented here will be useful for the on-going ginseng genome sequencing and further molecular-cytogenetic studies and breeding programs of ginseng.
Panax ginseng, FISH, 5SrDNA, 45S rDNA, DAPI band, Araliaceae
Ginseng (Panax ginseng C.A.Meyer, 1843) is highly valued owing to its medicinal properties (
Information regarding the chromosome number of ginseng has been available since 1936 (
The translocation of DNA blocks in some plants have been observed through cytogenetic investigations (e.g.
Probes labeled with different fluorophores for simultaneous detection have been widely employed in rDNA loci distribution analyses and dual-color FISH karyotyping (e.g.
Here, we used dual-color FISH to analyze the distribution of rDNA loci in Panax ginseng. In addition, we used the DAPI banding pattern to pair homologous chromosomes. Collectively, this made numbering of the chromosome of Panax ginseng possible for the first time. These data will be useful for future cytogenetic analyses and should enable a better understanding of the genomic history of ginseng, and can be used for subsequent distribution analyses of repeat sequences, retrotransposons, and chromosome-specific cytogenetic markers. Consequently, the results presented here will make a significant contribution to studies related to the on-going ginseng genome sequencing and the overall understanding of the Panax ginseng genome.
Stratified seeds of three ginseng cultivars ‘Sunun’, ‘Chunpoong’, ‘Gopoong’, and a local landrace ‘Hwangsook’ were provided by the Korea Ginseng Corporation (KGC) Natural Resources Research Institute (Daejeon, Korea). Stratified seeds were allowed to germinate in petri dishes with wet filter papers at 10–15°C. The root meristems were then excised (about 2 cm from the root tips), pretreated with 0.002M 8-hydroxyquinoline for 5 hours at 18°C, fixed in 90% acetic acid for 15 min at room temperature (RT, ~24°C), and then stored in 70% ethanol until use.
Somatic chromosome spreads were obtained using a modified version of the technique described by
A 9-kb fragment of 45S rDNA (18S-5.8S-25S) (
Slide pretreatment. To remove contaminating RNA, the slides were treated with RNase A buffer (RNase A final conc. 100 μg ml-1 in 2× SSC) for 1 hr at 37°C. The slides were then incubated in 0.01 M HCl for two minutes, followed by subsequent treatment in pepsin buffer [stock: 10% (w/v) pepsin in dH2O, working: 1:100 dilution in 0.01 M HCl] for 10 min at 37°C to lyse endogenous proteins that could cause background signals. Next, the chromosomes were fixed by treating the slides with 4% paraformaldehyde in 2× SSC. Finally, the slides were dehydrated in ethanol series (70%, 90%, 100%, 3 min each) and air-dried. The slides were washed in 2× SSC for 5 min (3×) between each step. All incubation steps at 37°C were conducted in a humidified chamber.
Probe hybridization.The hybridization mixture contained 50% formamide, 10% dextran sulfate, 2× SSC, 5 ng μl-1 salmon sperm DNA and 500 ng μl-1 of each probe DNA adjusted with DNase- and RNase-free water (Sigma, USA, #W4502) to a total volume of 40 μl/slide. The mixture was denatured at 90°C for 10 min and immediately kept on ice for at least 5 min prior to mounting on slides. After covering with a glass cover slip, the chromosomes were denatured at 80°C for 3–5 min on a hot plate. The slides were then immediately transferred into a humid chamber preset at 37°C and incubated overnight (~16 hr). The following day, the slides were washed in 2× SSC (15 min at RT), 0.1× SSC (35 min at 42°C), and finally 2×SSC (30 min at RT).
Signal detection. The slides were treated with TNB [0.1 M Tris-HCl, 0.15 M NaCl, 1% (w/v) blocking reagent] at RT for 5 min, after which they were subjected to antibody detection. Briefly, biotinylated 45S rDNA probe was detected with streptavidin-Cy3 conjugate (Zymed, USA), while digoxigenin-labeled 5S rDNA probe was detected using anti-digoxigenin-FITC conjugate (Sigma, USA). Both antibodies were diluted in TNB to a ratio of 1:100, and the slides were then incubated at 37°C for one hour. Excess reagents were subsequently washed off in TNT [0.1 M Tris-HCl, 0.15 M NaCl, 0.2% (v/v) Tween-20] at 37°C for 5 min (3×), after which they were subjected to dehydration in ethanol series (70%, 90%, 100%, 3 min each) and air-dried. Chromosomes were then counterstained with a premixed DAPI solution [1 μg ml-1 DAPI in Vectashield (Vector Laboratories, USA)].
Image capture and measurement. Well-spread chromosomes with well-preserved chromosome morphology were observed and captured using an Olympus BX51 fluorescence microscope equipped with a CCD camera (CoolSNAP™ cf) and filters for DAPI, FITC, and Cy3. The captured FISH images were analyzed, after which each homologue was measured 3–7 times using Genus™ version 3.1 (Applied Imaging, USA) to obtain the mean values. Raw images for each probe were saved separately and a pseudo-colored image of the merged signals was obtained for each chromosome spread. The sharpness value in Genus™ was set to 7 to enhance the details and texture of the chromosomes. Final images were edited using Adobe Photoshop CS3.
Chromosome numbering and pairing. Chromosome number assignment was based on the decreasing order of chromosome lengths, while homologous chromosome pairing was achieved according to the centromeric position (
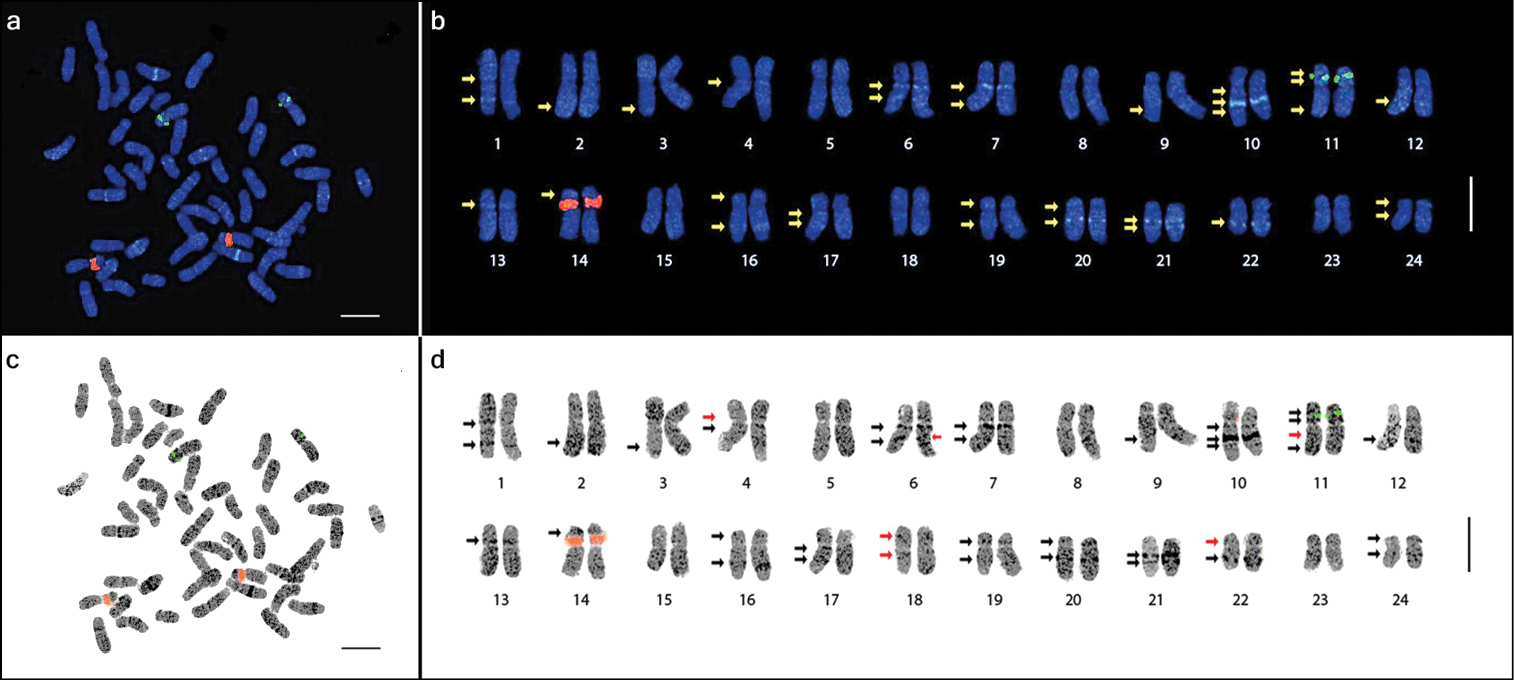
The three cultivars and one landrace of Panax ginseng evaluated in this study were all confirmed to have a chromosome complement of 2n=48 (Fig. 1). With reference to the centromere position (i.e. arm ratio), the complement comprised 12 metacentric (1–7, 11–13, 15, and 18), 9 submetacentric (8–10, 16–17, 19, and 22–24), and 3 subtelocentric (14 and 20–21) homologous pairs with a karyotype formula of 24m+18sm+6st. The chromosome lengths ranged from 3.27 to 6.30 μm (Table 1).
Chromosome complement of three Panax ginseng cultivars, ‘Sunun’ (a), ‘Gopoong’ (b), ‘Chunpoong’ (c), and one local landrace, ‘Hwangsook’ (d) showing 2n=48. One pair of 45S rDNA (red signals, yellow arrows) and one pair of 5S rDNA (green signals, white arrows) was observed among the four samples. Bar = 10 μm.
Chromosome analyses of Panax ginseng based on chromosome length and centromeric position.
| Chr. no. | Chr. length (μm) | Arm ratio (q/p) | Type | ||
|---|---|---|---|---|---|
| Short arm (p) | Long arm (q) | Total | |||
| 1 | 3.16 ± 0.12 | 3.17 ± 0.11 | 6.30 ± 0.22 | 1.002 | m |
| 2 | 2.64 ± 0.08 | 3.27 ± 0.12 | 6.05 ± 0.06 | 1.237 | m |
| 3 | 2.52 ± 0.23 | 3.61 ± 0.11 | 5.88 ± 0.18 | 1.434 | m |
| 4 | 2.54 ± 0.20 | 3.27 ± 0.22 | 5.64 ± 0.07 | 1.289 | m |
| 5 | 2.23 ± 0.27 | 3.35 ± 0.12 | 5.41 ± 0.17 | 1.506 | m |
| 6 | 2.05 ± 0.11 | 3.30 ± 0.06 | 5.31 ± 0.23 | 1.609 | m |
| 7 | 2.09 ± 0.07 | 3.35 ± 0.22 | 5.30 ± 0.14 | 1.605 | m |
| 8 | 1.54 ± 0.32 | 3.66 ± 0.13 | 5.23 ± 0.40 | 2.378 | sm |
| 9 | 1.52 ± 0.19 | 3.82 ± 0.13 | 5.08 ± 0.21 | 2.515 | sm |
| 10 | 1.77 ± 0.04 | 3.49 ± 0.06 | 5.04 ± 0.28 | 1.965 | sm |
| 11† | 2.13 ± 0.12 | 2.91 ± 0.13 | 4.94 ± 0.12 | 1.363 | m |
| 12 | 1.96 ± 0.07 | 3.03 ± 0.12 | 4.83 ± 0.28 | 1.547 | m |
| 13 | 2.04 ± 0.05 | 3.05 ± 0.04 | 4.82 ± 0.07 | 1.492 | m |
| 14‡ | 1.99§ ± 0.21 | 3.21 ± 0.14 | 4.80 ± 0.31 | 1.612| | st |
| 15 | 2.26 ± 0.17 | 2.58 ± 0.28 | 4.73 ± 0.49 | 1.143 | m |
| 16 | 1.55 ± 0.09 | 3.33 ± 0.10 | 4.72 ± 0.08 | 2.157 | sm |
| 17 | 1.59 ± 0.15 | 3.05 ± 0.07 | 4.50 ± 0.11 | 1.919 | sm |
| 18 | 2.09 ± 0.25 | 2.54 ± 0.19 | 4.50 ± 0.06 | 1.214 | m |
| 19 | 1.39 ± 0.12 | 2.78 ± 0.17 | 4.11 ± 0.21 | 1.998 | sm |
| 20 | 1.05 ± 0.04 | 3.24 ± 0.07 | 4.09 ± 0.06 | 3.067 | st |
| 21 | 0.90 ± 0.05 | 3.02 ± 0.21 | 3.80 ± 0.13 | 3.355 | st |
| 22 | 1.32 ± 0.06 | 2.32 ± 0.10 | 3.56 ± 0.09 | 1.761 | sm |
| 23 | 1.25 ± 0.11 | 2.30 ± 0.22 | 3.38 ± 0.09 | 1.836 | sm |
| 24 | 1.13 ± 0.25 | 2.08 ± 0.24 | 3.27 ± 0.10 | 1.840 | sm |
†5S rDNA, ‡45S rDNA, §satellite length, |value obtained using satellite instead of short arm, m: metacentric, sm: submetacentric, st: subtelocentric (
Only one pair of satellited chromosomes (pair 14) was observed, and the only locus of 45S rDNA in the genome was localized at the secondary constriction of this subtelocentric chromosome (Figs 2, 3 and Table 2). Moreover, one locus of 5S rDNA signal was detected at the intercalary region of the short arm of chromosome 11. This locus was flanked by two DAPI bands. There was no variation in the number of rDNA loci among the three cultivars and one landrace of Panax ginseng investigated in this study (Fig. 1).
Metaphase spread of Panax ginseng 2n=48 chromosomes (a and c) and the karyotype idiogram showing 24 homologous pairs (enlarged; b and d) arranged in decreasing lengths. The 5S and 45S rDNA loci are shown as green and red signals, respectively. DAPI bands (arrows) were detected in various intensities and inverse images (c and d) were obtained to emphasize these DAPI bands. Note the heterochromatic dots (dark dots in d). The red arrows in d indicate the six bands observed after inversing the image. Bar=5 μm.
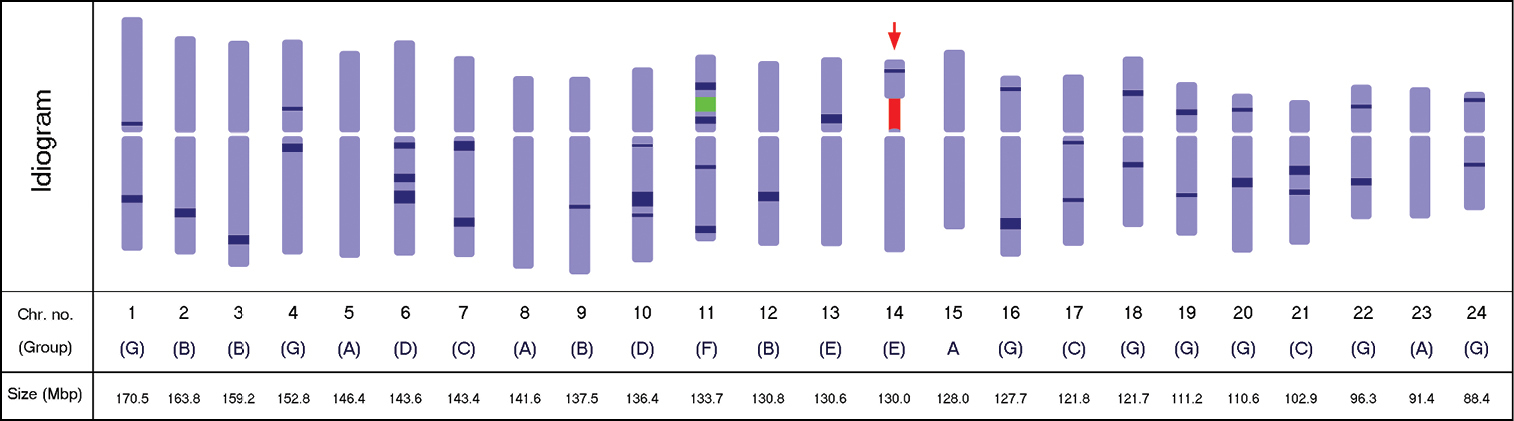
Diagrammatic idiogram of the Panax ginseng karyotype showing the 5S (green) and the 45S (red) rDNA loci, and the 38 observed DAPI bands (dark blue), 12 on the short arm and 26 on the long arm. The satellited chromosome is indicated by the red arrow. DAPI band depths indicate relative intensities. Chromosomes were grouped according to the DAPI band pattern on each arm. The estimated relative size of each chromosome is presented in mega base-pairs.
Numerous DAPI-binding heterochromatic regions were dispersed along all chromosomes and were visible as DAPI dots. These dots, similar to those in chromosomes 5 and 8, did not form distinct DAPI bands. Both the DAPI dots and bands were made more easily visible by inverting the images (Fig. 2c and d).
In addition to the rDNA loci, the presence of several observable DAPI bands along the chromosome complement made identification of homologous pairs possible. The number of the observed bands further increased as the resolution increased after subsequent enhancement of the image sharpness in Genus™. A total of 32 DAPI bands were initially observed in the sharpness-enhanced DAPI images, but six additional DAPI bands were observed after using the inverse tool of Genus™ with adjustment to the brightness and contrast, resulting in a total of 38 bands (Fig. 2b and d).
Twelve of the observed DAPI bands were localized on the short arms, while 26 were on the long arms (Table 2). Among the 24 chromosomes, four had no band (5, 8, 15, and 23), six had one band (2, 3, 9, and 12–14), 11 had two bands (1, 4, 7, 16–22, 24), two had three bands (6 and 10), and one had four bands (11). Furthermore, chromosomes were grouped according to the presence or absence of DAPI bands on each arm (Fig. 3). Group A had no band on either arm (pairs 5, 8, 15, and 23), group B had no band on the short arm, but one band on the long arm (2, 3, 9, and 12), group C had no band on the short arm, but two bands on the long arm (7, 17, and 21), group D had no band on the short arm, but three bands on the long arm (6 and 10), group E had one band on the short arm, but none on the long arm (13 and 14), group F had two bands on both arms (11), and group G had one band on each arm (1, 4, 16, 18–20, 22, and 24).
Summary of the rDNA and DAPI band distribution patterns.
| Chr. no. | rDNA distribution | DAPI band distribution | Remarks | ||
|---|---|---|---|---|---|
| 5S | 45S | Short arm | Long arm | ||
| 1 | - | - | 1 | 1 | Pericentric on short arm, more intense intercalary on long arm |
| 2 | - | - | - | 1 | Dispersed, weak, subtelomeric |
| 3 | - | - | - | 1 | Subtelomeric, average intensity |
| 4 | - | - | 1 | 1 | Pericentric on both arms. Weaker on short arm |
| 5 | - | - | - | - | |
| 6 | - | - | - | 3 | One intense pericentric, two intercalary with weaker proximal |
| 7 | - | - | - | 2 | intense pericentric, weak distal |
| 8 | - | - | - | - | |
| 9 | - | - | - | 1 | Weak, intercalary |
| 10 | - | - | - | 3 | Weak pericentric, two intercalary with very intense middle and weak distal |
| 11 | 1 | - | 2 | 2 | Two moderate intensity flanking 5S rDNA on short arm, one weak intercalary and one weak subtelomeric on long arm. 5S rDNA moderate intensity |
| 12 | - | - | - | 1 | Intercalary, moderate intensity |
| 13 | - | - | 1 | - | Pericentric, weak |
| 14† | - | 1 | 1 | - | Subtelomeric on satellite, weak; intense 45S rDNA |
| 15 | - | - | - | - | |
| 16 | - | - | 1 | 1 | Weak subtelomeric on short arm, more intense intercalary on long arm |
| 17 | - | - | - | 2 | Weak pericentric, weak intercalary |
| 18 | - | - | 1 | 1 | Weak intercalary on short arm, weak pericentric on long arm |
| 19 | - | - | 1 | 1 | Intercalary on both arms, more intense on short arm |
| 20 | - | - | 1 | 1 | Intercalary on both arms, more intense on long arm |
| 21 | - | - | - | 2 | Intercalary, proximal more intense than distal |
| 22 | - | - | 1 | 1 | Intercalary on both arms, more intense on long arm, long arm signal more intense than that on chromosome 20 long arm |
| 23 | - | - | - | - | |
| 24 | - | - | 1 | 1 | Weak subtelomeric on short arm, more intense intercalary on long arm |
| Total | 1 | 1 | 12 | 26 | |
†satellited chromosome
In addition to the chromosome length, centromeric position, and rDNA loci distribution, we utilized the observed DAPI bands to characterize the chromosomes. Collectively, these DAPI bands could be very useful in identifying homologues for further cytogenetic analyses, especially of the Panax ginseng genome, which comprises a large number of chromosomes with mostly similar sizes. The distinguishing features of each chromosome are presented in Table 2.
There is currently not much genomic or cytogenetic information available for ginseng. Consequently, there are no established cytogenetic markers for the identification of homologous chromosomes. This lack of data has limited our understanding of the karyotype of ginseng and therefore its phylogenetic relationship with other species in the genus Panax. In this study, we exploited the usefulness of the 5S and 45S rDNA and the DAPI-binding heterochromatins as molecular cytogenetic markers in pairing homologous chromosomes by analyzing their distribution in the Panax ginseng genome.
We detected only one locus each for 5S and 45S rDNA, which is in agreement with the results reported by
Localization of the rDNA resulted in our only being able to easily pair two out of the 24 homologues. However, the existence of several DAPI bands distributed along most of the chromosomes greatly facilitated the identification of the other homologous pairs, which otherwise would have been challenging owing to the very low size diffe-rence among most ginseng chromosomes. As a result, DAPI banding, which has been utilized in several previously conducted studies (e.g.
Chromosomal DAPI bands are caused by the preferential binding of DAPI to AT-rich heterochromatic DNA segments (
The use of the rDNA loci number and distribution pattern of other Panax species can be useful in deducing the phylogenetic relationship among these species.
Karyotype data are essential to understanding the phylogenetic relationships among species belonging to the same family (
Most species belonging to the family Araliaceae are 2n=24 or 2n=48, except for a few genera that have little chromosomal number variation (
Considering a basic chromosome number of 12 or 6, ginseng would be considered a tetraploid or octoploid, respectively; the latter having a more ancient nature. Recently,
Our data showed a somatic cell chromosome complement of 2n=48, supporting previously reported chromosome numbers (
Additionally, based on localization of the 45S rDNA near the centromere area and the intercalary position of the 5S rDNA, it is just as likely that these loci were favored to survive locus loss from non-additive recombination over their duplicated counterparts, which probably would have been in more distal positions, or epigenetically silenced (
The first report of Panax ginseng karyotype using ribosomal DNA and DAPI bands as cytogenetic markers is presented here. The presence of long stretches of DAPI-binding heterochromatin was useful in the detailed karyotyping. The results presented here will be useful in further cytogenetic analyses and the on-going genome sequencing of ginseng. More cytogenetic research is needed to understand the cytogenetic history of ginseng and other species in the genus Panax. Further comparative cytogenetic analyses among its close relatives will provide more insight, and further genomic analyses of the heterochromatin distribution will enhance our knowledge of its genomic history.
This work was supported by the Next-Generation BioGreen21 Program (No.PJ008202), Rural Development Administration, Republic of Korea, and by the Sahmyook University Research Fund (No. RI2011039).