|
||
|
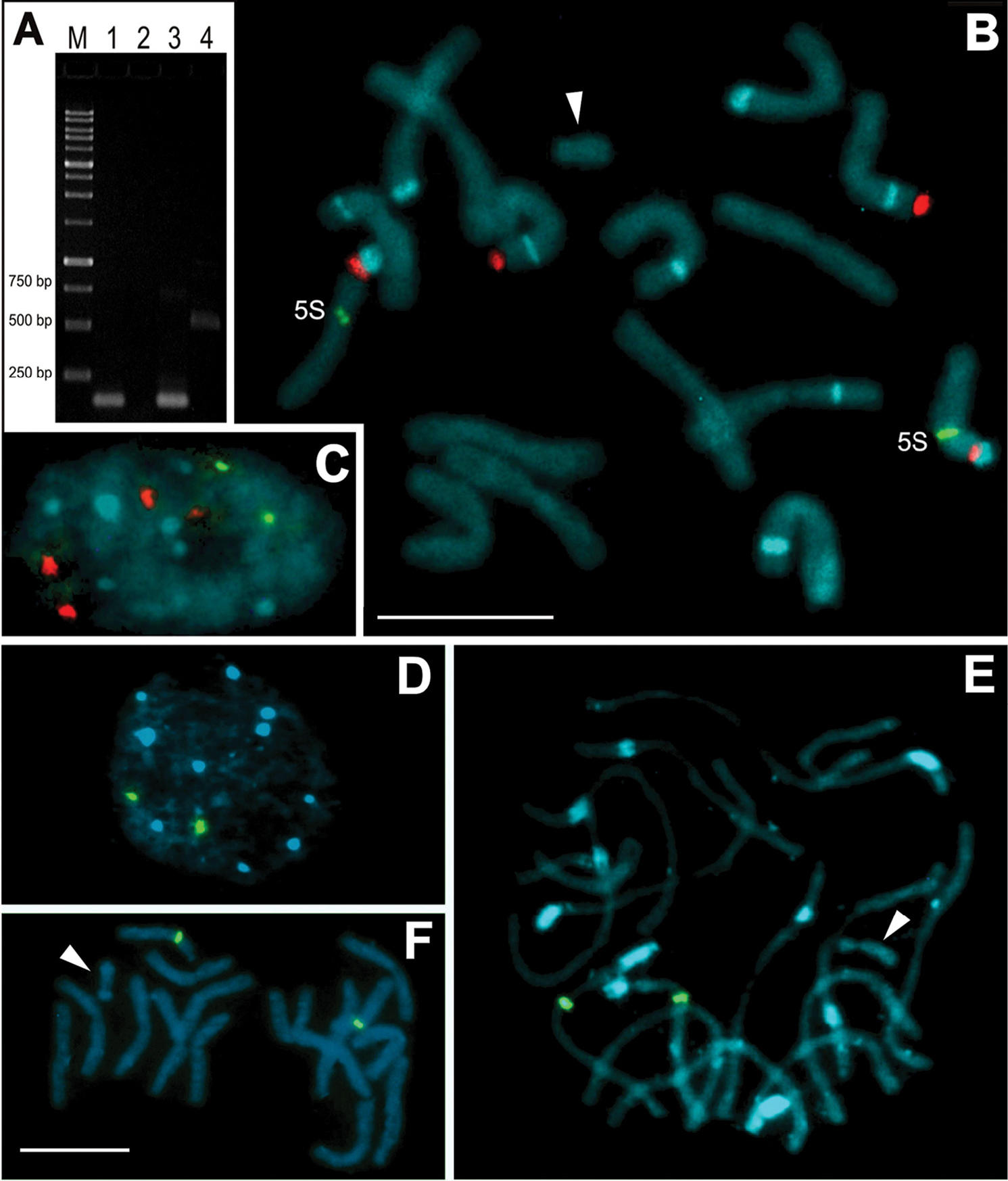
Detection of 5S rDNA by PCR and FISH using probes produced with genomic and B chromosome DNA templates. A Electrophoresis gel containing: M = ladder with 250, 500, 750, 1000 bp. Lanes 1 and 3 represent fragments of about 120 bp using the UP primers using the microdissected B chromosome as template (1) and the genome as template (3). Lanes 2 and 4 represent the PCR using 5S-plant primers with microdissected B chromosome as template (see the absence of fragment in lane 2) and with genome as template (see a fragment with about 500 bp length in lane 4 B–C Double FISH showing four hybridization sites for 35S rDNA using the pTa71 probe (red) and two sites for 5S rDNA using the UP primer probe of the genome (green) in the chromosomes (B) and nucleus (C) of C. strigilatum D–F FISH using the 5S rDNA fragment amplified, using the UP primers and the microdissected B DNA as template. Note only two signals in the nucleus and prometaphase chromosomes of C. strigilatum (D, E respectively). Note also the absence of hybridization signals in the Bs (arrowheads). Bands in blue color represent AT-rich regions identified by DAPI staining. Bar = 10 µm. |