|
||
|
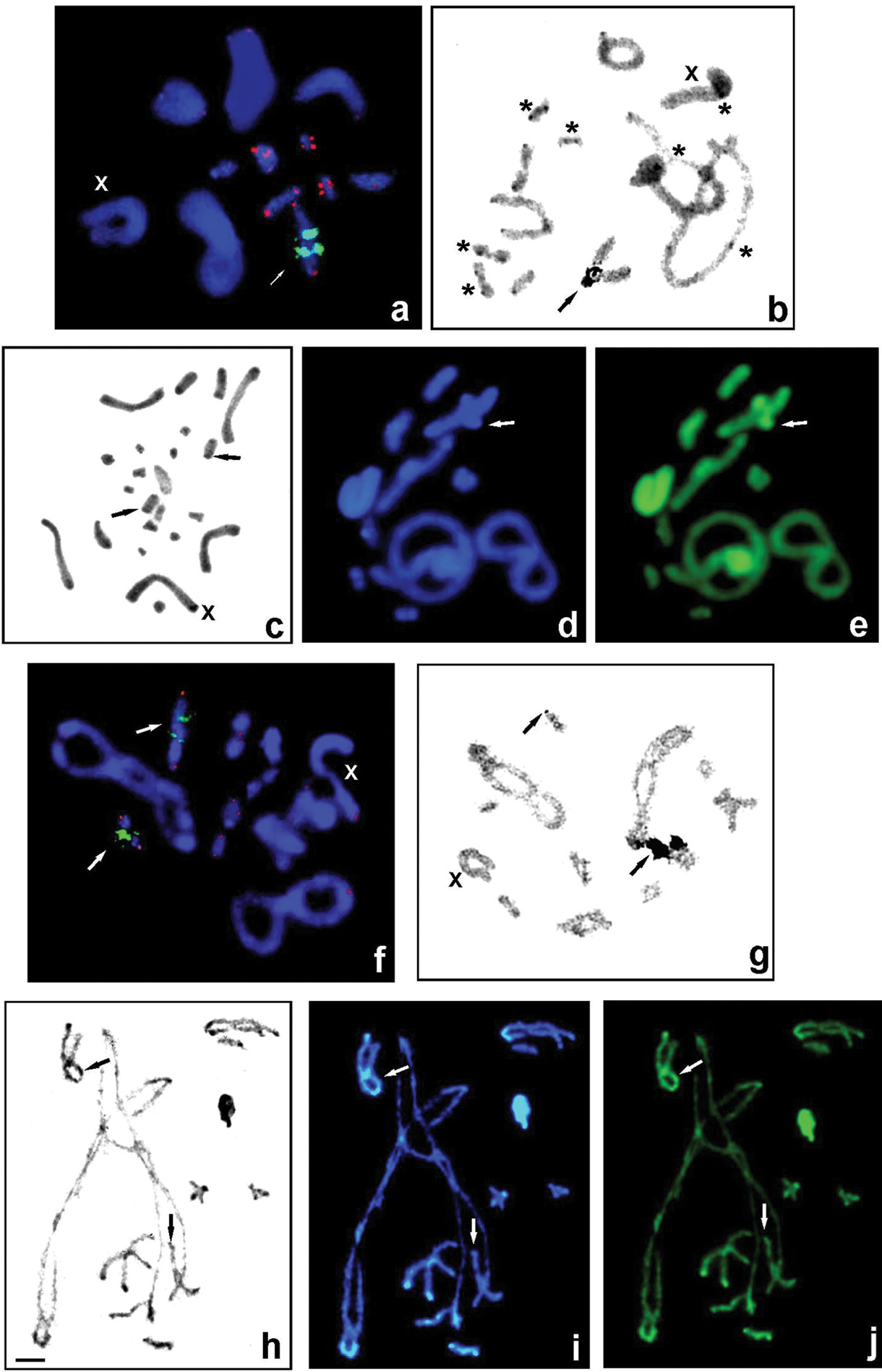
Gampsocleis species with 2n = 23 chromosomes (male): G. glabra (a–e) and G. abbreviata (f–j) studied using different techniques: FISH using 18S rDNA (green) and telomeric TTAGG (red) probes (a, f) and silver staining in diakinesis (b,g), C-banding of spermatogonial metaphase (c) and diplotene (h), and fluorochrome staining of heterochromatin with DAPI (blue) and CMA3 (green) (d, i, e, j). Arrows indicate rDNA clusters located near the telomeric region on the 5th bivalent (a, f) and in a telomeric position on the short bivalent (f); active NORs co-localized with rDNA (b, g, black arrows); thin C-bands (c, h, black arrows) and weak DAPI+ (d, i, white arrows) and bright CMA3+ signals located near the telomeric region on the medium-sized bivalent (e, j, white arrows) as well DAPI-/CMA3+ signals on the telomeric region of the short bivalent (i,j, white arrows). Bi-armed chromosomes are marked by asterisks (b). X, sex chromosome. Scale bar: 10 µm. |