|
||
|
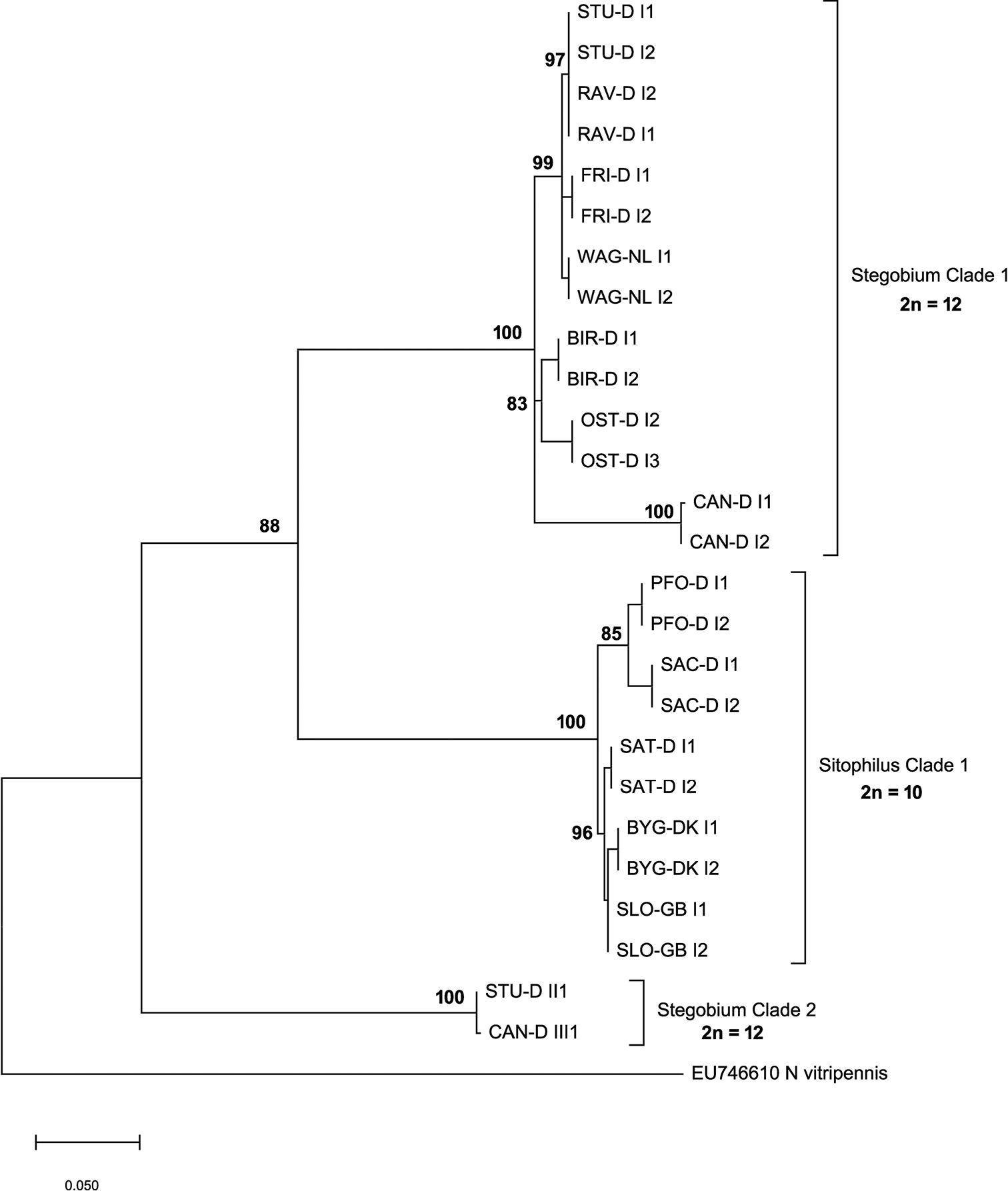
Evolutionary relationships of different strains of L. distinguendus based on a partial COI fragment. The evolutionary history was inferred by using the Maximum Likelihood method and Hasegawa-Kishino-Yano model (Hasegawa et al. 1985). The tree with the highest log likelihood (-2312.56) is shown. Percentages of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to the branches. The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 65.23% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. |