(C) 2011 María Georgina Poggio. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
In the present work, we analysed the male meiosis, the content and distribution of heterochromatin and the number and location of nucleolus organizing regions in Microtomus lunifer (Berg, 1900) by means of standard technique, C- and fluorescent bandings, and fluorescent in situ hybridization with an 18S rDNA probe. This species is the second one cytogenetically analysed within the Hammacerinae. Its male diploid chromosome number is 31 (2n=28+X1X2Y), including a minute pair of m-chromosomes. The diploid autosomal number and the presence of m-chromosomes are similar to those reported in Microtomus conspicillaris (Drury, 1782) (2n=28+XY). However, Microtomus lunifer has a multiple sex chromosome system X1X2Y (male) that could have originated by fragmentation of the ancestral X chromosome. Taking into account that Microtomus conspicillaris and Microtomus lunifer are the only two species within Reduviidae that possess m-chromosomes, the presence of this pair could be a synapomorphy for the species of this genus. C- and fluorescent bandings showed that the amount of heterochromatin in Microtomus lunifer was small, and only a small CMA3 bright band was observed in the largest autosomal pair at one terminal region. FISH with the 18S rDNA probe demonstrated that ribosomal genes were terminally placed on the largest autosomal pair. Our present results led us to propose that the location of rDNA genes could be associated with variants of the sex chromosome systems in relation with a kind of the sex chromosome systems within this family. Furthermore, the terminal location of NOR in the largest autosomal pair allowed us to use it as a chromosome marker and, thus, to infer that the kinetic activity of both ends is not a random process, and there is an inversion of this activity.
Hemiptera, Reduviidae, Hammacerinae, meiosis, m-chromosomes, evolutionary trends, rDNA-FISH
Reduviidae is the largest family of predaceous land Hemiptera
and includes about 6500 species and subspecies in 930 genera and 22
subfamilies. These insects are abundant, occur worldwide, and are
voracious predators (thus their name, “assassin bugs”) (
All hemipteran species possess holokinetic chromosomes,
i.e. chromosomes without primary constrictions and, hence, without
localized centromeres. This order is unique in that the autosomes,
m-chromosomes and sex chromosomes have different meiotic behaviours.
During mitosis microtubules attach to the entire length of sister
chromatids, and at anaphase they segregate parallel to each other and
perpendicular to the polar spindle (holokinetic behaviour) (
As a rule, autosomal bivalents are chiasmatic, whereas sex chromosomes and m-chromosomes are achiasmatic (
Apart from the general characteristics of hemipteran species previously described, the Reduviidae
are characterized by a modal diploid number of autosomes of 20 with a
range between 10 and 34, and both simple and multiple sex chromosome
systems (XY/XX, X0/XX, and XnY/XnXn; male/female) (
Furthemore, cytogenetic data for species belonging to Reduviidae
point to the presence of C-heterochromatin at terminal regions on a few
or all autosomal pairs, and/or on one of the sex chromosomes, whereas
the other one is completely heterochromatic (
So far, the location of nucleolus organizing regions
(NORs) has been analysed in only 14 reduviid species by Ag-NOR,
fluorescent banding and/or fluorescent in situ hybridization (FISH) with ribosomal DNA (rDNA) probes (18S, 26S or 45S). These results show that in Reduviidae
the NOR can be located either at terminal position on one autosomal
pair, or on the sex chromosomes. The presence of NORs in both X and Y
chromosomes was reported in two species belonging to two different
subfamilies (Harpactorinae and Triatominae) (
In the present work, we analysed in detail the male meiosis of Microtomus lunifer (Berg, 1900) (Hammacerinae) to verify the presence of a pair of m-chromosomes, the content and distribution of heterochromatin by C- and fluorescent bandings, and examined the number and location of NORs by FISH. Lastly, the position of a NOR at the terminal region of the largest autosomal pair allowed us to use it as a chromosome marker and to describe its behaviour during both meiotic divisions.
Material and methods InsectsWe used three males of Microtomus lunifer from Pampa del Indio, Chaco province (coll. 2008).
Chromosome preparationsAll the analysed specimens were brought alive to the
laboratory. The male gonads were dissected in physiological solution.
Afterwards, one of the testes was fixed for 15–30 min in freshly
prepared Carnoy fixative (ethanol: chloroform: acetic acid, 6:3:1),
and was kept at 4ºC in 70% ethanol for meiotic studies. Slides were
prepared by the squash technique in a drop of 2% iron acetic
haematoxylin following conventional procedures. For C- and fluorescent
bandings, and FISH techniques, spread chromosome preparations were
made from the other testis as described in
C- and fluorescent bandings were then applied to
spread chromosome preparations to reveal heterochromatin and its base
composition. C-banding was performed according to
Fluorescent banding with AT-specific DAPI and
GC-specific chromomycin A3 (CMA3; Fluka BioChemika) was carried out as
follows: after removal from freezer, the slides were placed immediately
into cold 70% ethanol for 2 min. Then, they were transferred through
80% and 100% ethanol, 30 sec each, and air-dried. The slides were
submerged in a coplin jar with methanol for two hours. Once dried, they
were rinsed with Mc Ilvaine´s buffer pH 7 (0.1 M citric acid, 0.2 M
Na2HPO4, in distilled water). Each chromosome preparation was dyed with
75 µl of DAPI solution (0.01 mg/ml, in Mc Ilvaine’s buffer), covered
with 24x50 mm transparency cover slides, and kept at room temperature
(RT) for 20 min in darkness in a moist chamber. Afterwards, the
preparations were rinsed three times using distilled water, Mc
Ilvaine’s buffer and distilled water. Then, the slides were dyed with
50 µl of CMA3 solution (0.6 mg/ml, in Mc Ilvaine´s buffer), covered
with 24x50 mm transparency cover slide, and incubated at RT for 1 hour
in dark in a moist chamber. After this period, the preparations were
rinsed again with distilled water, Mc Ilvaine’s buffer and distilled
water, and then let them air-dried. The slides were mounted in Antifade
based on DABCO (Sigma Aldrich; for composition see
Unlabelled 18S rDNA probes were generated by polymerase
chain reaction (PCR) using universal arthropod primers: forward
5‘-CCTGAGAAACGGCTACCACATC-3’ and reverse 5‘-GAGTCTCGTTCGTTATCGGA-3’ (
The location of NOR regions in the largest autosomal pair of Microtomus lunifer allowed us to analyse the behaviour of the terminal regions which were kinetically active. The number of cells at metaphase I and metaphase II, in which the kinetically active terminal regions of this autosomal pair were associated to the NOR (Figs 5d, g;) or not (Figs 5e, f), were counted. The hypotheses described below were tested using a Chi-squared goodness of fit test.
H01: the kinetic activity of both ends (with/without NOR) at both meiotic divisions is a random process.
H02: the chromosome end that is active during the first meiotic division becomes inactive during the second one and vice versa.
Microscopy and image processingPreparations were observed in a Leica DMLB microscope equipped with a Leica DFC350 FX CCD camera and Leica IM50 software, version 4.0 (Leica Microsystems Imaging Solutions Ltd., Cambridge, UK). Black-and-white images of chromosomes were recorded separately for each fluorescent dye. Images were pseudocoloured (light blue for DAPI, green for CMA3, red for Cy3) and processed with an appropriate software.
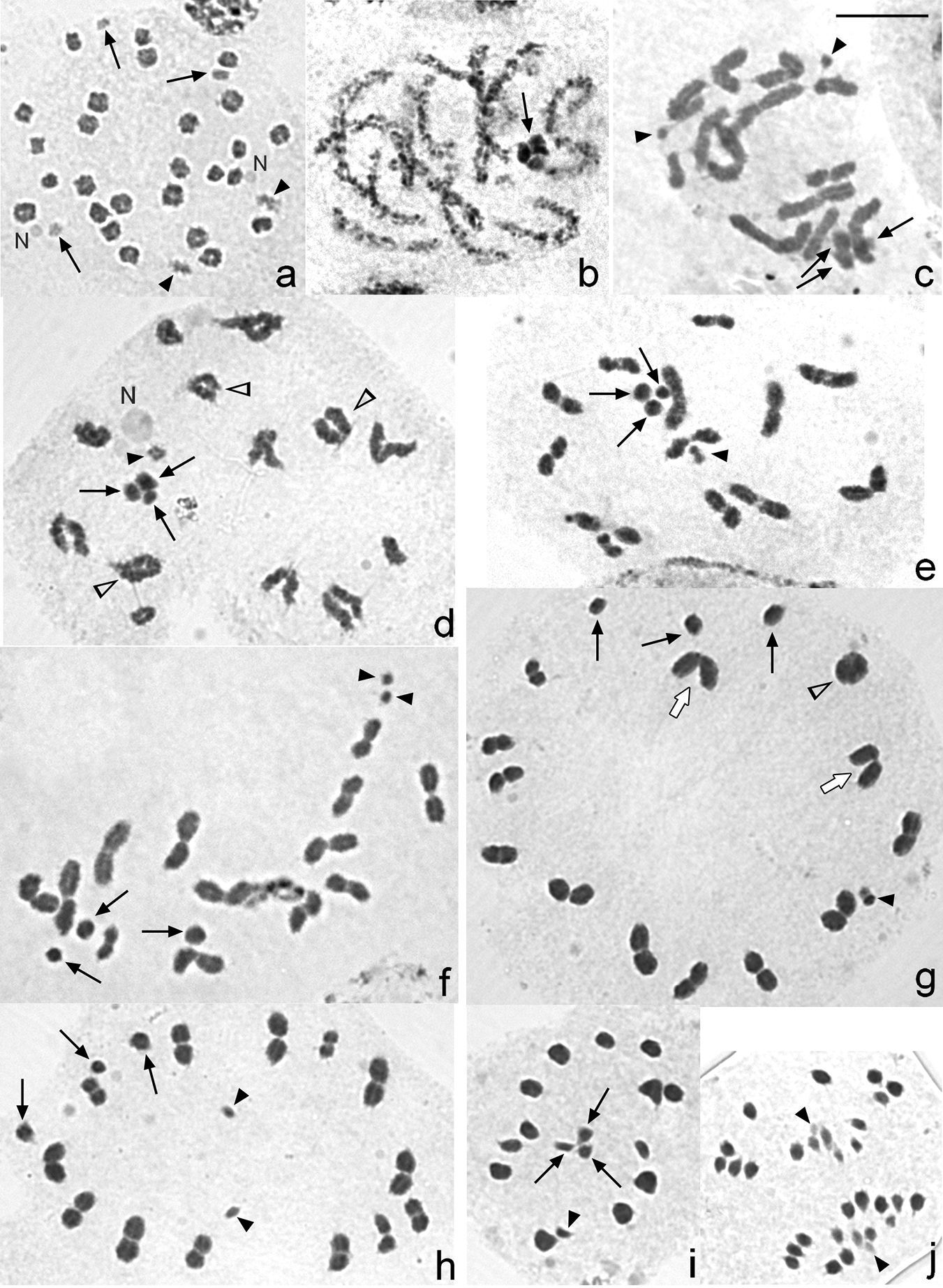
Results Male chromosome complement and meiosisMicrotomus lunifer possesses a male diploid chromosome number of 31, and its complement comprises 14 autosomal bivalents and a multiple sex chromosome system X1X2Y (Fig. 1). In spermatogonial prometaphase, the sex chromosomes and an autosomal pair are easily recognized because of their small size, whereas the rest of the autosomes cannot be distinguished due to their similar size. An association between a nucleolus and an autosomal pair is observed (Fig. 2a).
At early pachytene, it is not possible to individualize each autosomal bivalent. However, the three sex chromosomes are positively heteropycnotic and lie close to each other forming a pseudo-trivalent. At late pachytene the bivalents continue their condensation, and the sex chromosomes become isopycnotic (Fig. 2b). From diplotene onwards, 13 autosomal bivalents, two univalents and three sex chromosomes are clearly distinguished in some cells (Fig. 2c, f), whereas in other ones 14 autosomal bivalents and three sex chromosomes are also observed (Fig. 2d, e). It can be noticeably seen that the sex chromosomes differ slightly in size (Fig. 2d–f). At metaphase I, the sex univalents lie at the periphery of the ring formed by the autosomal bivalents, and their different size is evident (Fig. 2g, h). At this stage, the smallest chromosome pair does not show any defined position and can be found either being part of the ring (Fig. 2g) or at its centre (Fig. 2h). This smallest pair can be observed migrating precociously in some cells (33 out of 100 cells) (Fig. 2h). At anaphase I, autosomal bivalents divide reductionally, while the sex chromosomes segregate equationally. Therefore, at telophase I two nuclei with 17 chromosomes each (14A+X1X2Y) are observed. Second meiotic division follows immediately after telophase I without an interkinesis stage. At metaphase II, the autosomes dispose at the equatorial plane forming a ring, and in the centre of it the sex chromosomes form a pseudo-trivalent (Fig. 2i). The Y chromosome is orientated towards the spindle pole opposite to that of X1 and X2. At anaphase II, 15 chromosomes migrate to one of the poles (14A+Y) and 16 to the opposite one (14A+X1X2) (Fig. 2j).
There is usually only one chiasma on each autosomal bivalent, which can be terminally or, less frequently, subterminally located, although they can show two chiasmata (Fig. 2d, g). Cells with two ring bivalents are seldom observed, while those with three ring bivalents are even rarer (Fig. 2d). In this species three kinds of bivalents are observed: rod (Fig. 2c–h), ring (Fig. 2d, g) and V-shaped (Fig. 2g) bivalents from diplotene to metaphase I. Mean chiasma frequency in cells at diakinesis-metaphase I is 14.76, being 15 (38.7%, 93 analysed cells) and 14 (68.7%, 99 analysed cells) the most frequent number of chiasmata at diakinesis and at metaphase I, respectively.
Male meiotic karyotype of Microtomus lunifer. Chromosomes are counterstained with DAPI; the largest autosomal pair is recognized by the presence of the rDNA hybridization signals. Bar = 10 μm
Male meiosis in Microtomus lunifer. a Spermatogonial prometaphase b Late pachytene c Diplotene. The smallest chromosome pair is observed as two univalents (black arrowheads) d Diplotene. The smallest chromosome pair is as a pseudo-bivalent (black arrowhead) e Diakinesis. The smallest chromosome pair is as a pseudo-bivalent (black arrowhead) f Diakinesis. The smallest chromosome pair is observed as two univalents (black arrowheads) g Metaphase I. The smallest chromosome pair (black arrowhead) formed a pseudo-bivalent and is placed in the bivalent autosomal ring h Metaphase I. The smallest chromosome pair lies in the centre of the ring and migrates precociously (black arrowheads) i Metaphase II j Anaphase II. Black arrows: sex chromosomes. Black arrowheads: smallest chromosome pair. White arrowheads: autosomal bivalents with two chiasmata. White arrows: V-shaped bivalents. N: nucleolus. Chromosomes are stained with 2% iron acetic haematoxylin. Bar = 10 µm
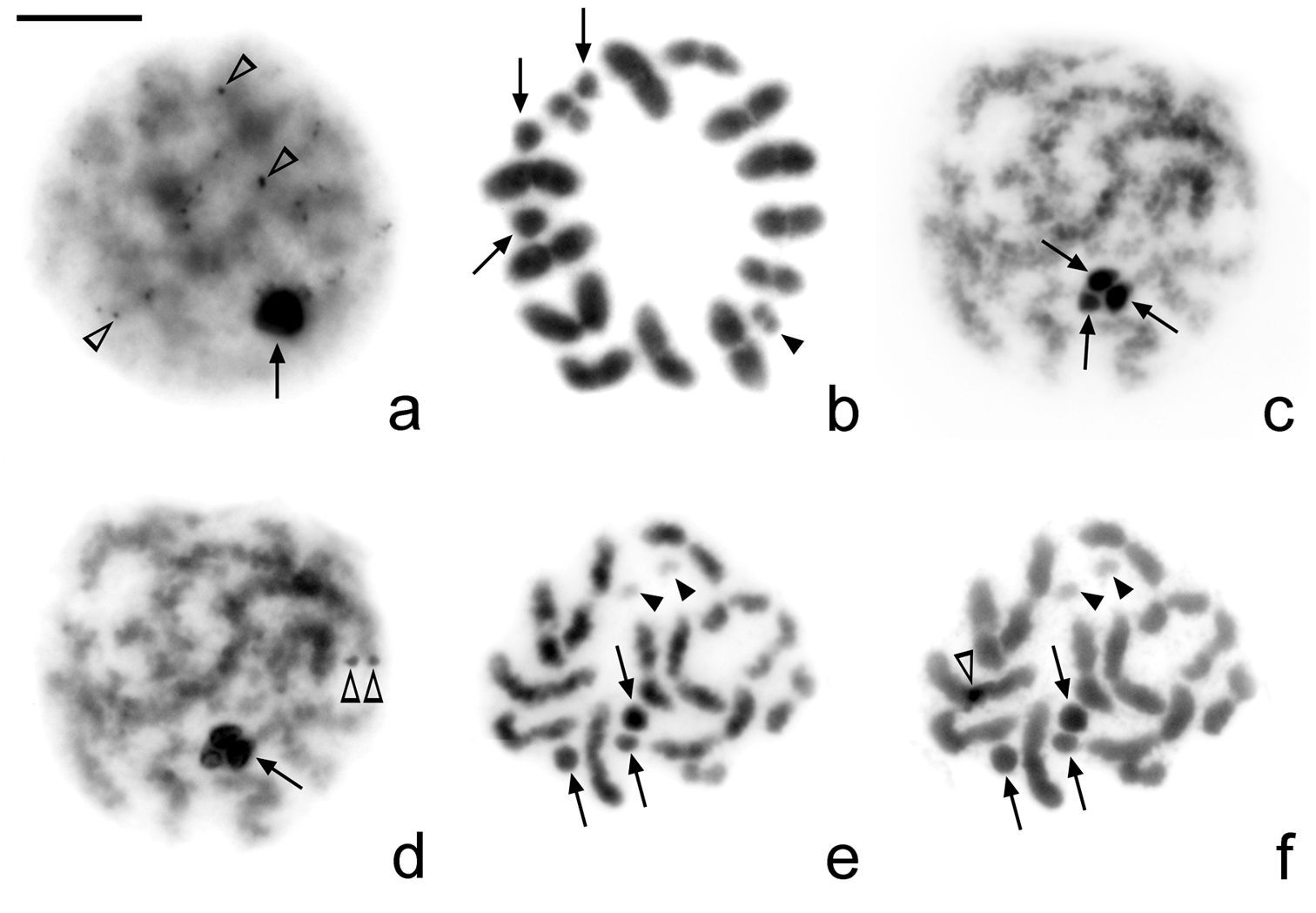
The amount of heterochromatin in Microtomus lunifer is small: very small C-positive dots (from 10 to 20) are detected in cells at leptotene-zygotene. At this stage, the sex chromosomes are observed as completely C-positive (Fig. 3a). However, this C-banding pattern can no longer be detected from diplotene onwards (Fig. 3b). All meiotic chromosomes show uniform staining with DAPI (Fig. 3c, e) and CMA3 fluorochromes (Fig. 3d, f), except for the largest autosomal bivalent. A small CMA3 bright band is observed at one of the terminal regions of the largest autosomal pair (Fig. 3d, f). Besides, the smallest pair of chromosomes is both DAPI and CMA3 dull.
Microtomus lunifer. a–b C-banding and c–f Fluorescent banding: c, e DAPI and d, f CMA3. a Leptotene-zygotene. Very small C-positive dots can be observed in the autosomal chromatin; sex chromosomes are C-positive b Metaphase I. No C-positive bands can be detected c–d Pachytene e–f Diakinesis. No DAPI (c, e)and neither CMA3-positive bands (d, f) can be detected, except for a small CMA3 bright band in one of the terminal regions of the largest autosomal pair. Arrows: sex chromosomes. Black arrowheads: smallest autosomal pair. White arrowheads: positive dots/bands. Bar = 10 µm
In Microtomus lunifer, FISH experiments with 18S rDNA probes reveal a single cluster placed at one terminal region of the largest autosomal pair (Fig. 4a–g). In spermatogonial metaphases, it is clearly observed that the hybridization signals are at terminal regions of both sister chromatids of both homologous chromosomes (Fig. 4a). From diplotene onwards, the hybridization signals are detected at one terminal region of the largest autosomal bivalent (Fig. 4b, c). However, both at metaphase I and metaphase II the NOR-autosomal pair shows two different orientations depending on the location of the hybridization signals: the ends with the NOR oriented to the poles (Fig. 4d, g) or the ends without NOR oriented to the poles (Fig. 4e, f).
Microtomus lunifer. Fluorescent in situ hybridization with an 18S rDNA probe. a Spermatogonial prometaphase b Diplotene c Diakinesis d–e Metaphase I f–g Metaphase II. Hybridization signals in red. Chromosomes are counterstained with DAPI (blue). Arrows: sex chromosomes. White arrowheads: smallest autosomal pair. Bar = 10 µm
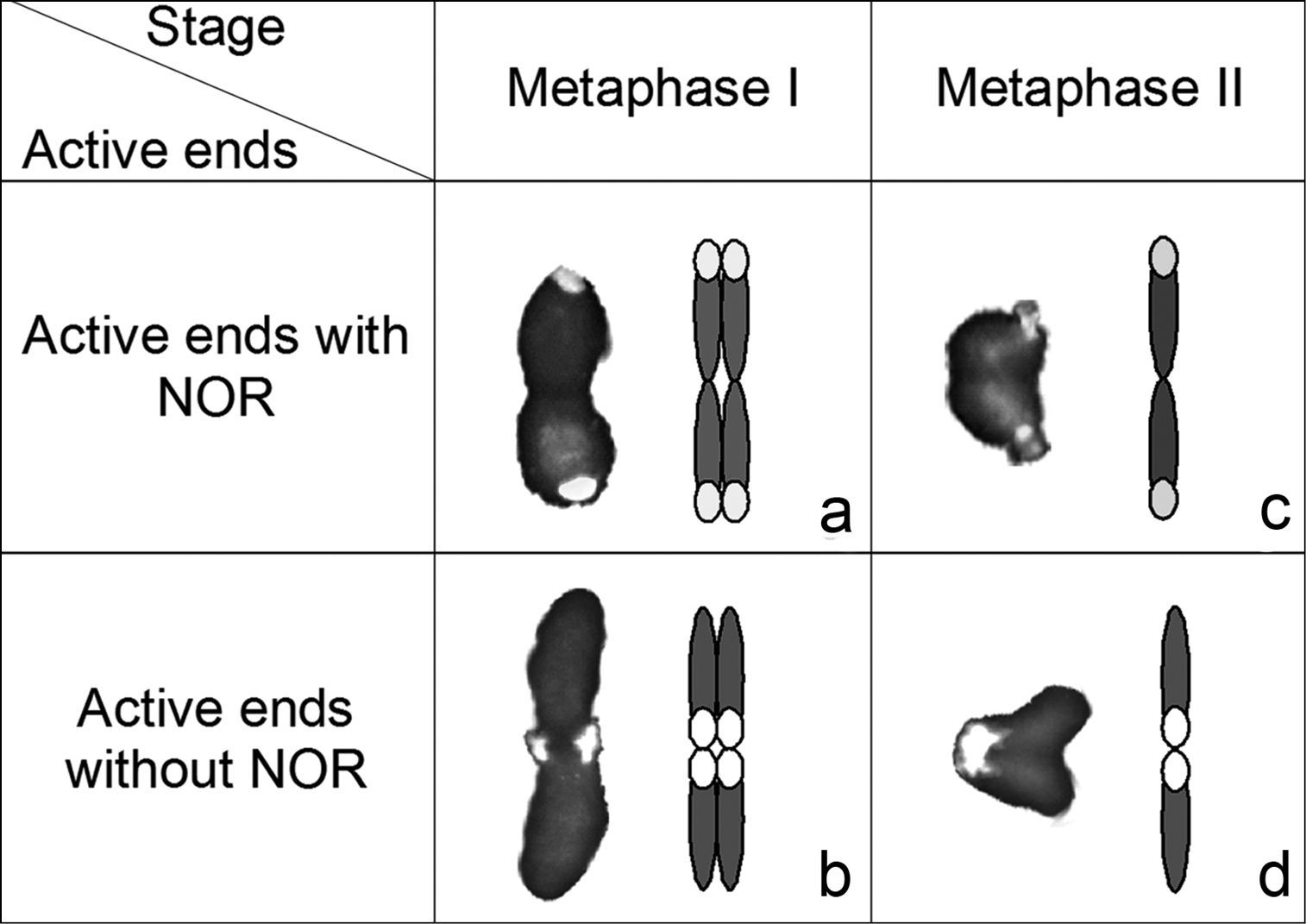
In Microtomus lunifer, FISH experiments provide a reliable chromosome marker in the NOR-autosome pair to analyse its meiotic behaviour during both meiotic divisions (Figs 4, 5). The presence of a single cluster of rDNA at only one of the ends of each homologous chromosome of the NOR-bivalent allows us to distinguish whether both ends (carrying the NOR or not) take part in the kinetic behaviour of this autosomal pair. At metaphase I, this NOR-bivalent is axially oriented and shows two types of configuration: either the chromosome ends bearing the hybridization signals (Figs 4d, 5a) or the ends that do not bear them are directed towards the poles (Figs 4e, 5b). At metaphase II, the sister chromatids reach an axial orientation and present the same two arrangements: either the chromatid ends carrying the hybridization signals (Figs 4g, 5c) or the other ends that do not carry those (Figs 4f, 5d) are oriented towards the poles.
To test whether the kinetic activity of both ends is a random process at metaphase I and metaphase II, the configurations of the NOR-autosomal bivalent in three individuals were scored. The results demonstrate that at metaphase I the kinetic activity of this NOR-bivalent is restricted to the chromosome ends that do not carry the hybridization signals in 67% of the analysed cells (216 out of 322), whereas in the remaining 33% the kinetic activity occurs at the ends that carry the hybridization signals (Table 1). At metaphase II, however, the kinetic activity is located at the chromatid ends bearing the NOR in 76% of the cells (117 out of 154), and in the remaining cells, at the chromatid ends without it (24% of the cells). Comparing the frequencies of configurations of this NOR-autosome pair between cells at metaphase I and metaphase II, we can observe similar frequencies of cells in which the kinetic activity at metaphase I is restricted to the chromosome ends not carrying the NOR and cells at metaphase II where the kinetic activity is located at the chromatid ends bearing the NOR, and vice versa. Statistical analysis corroborates that: i) the kinetic activity of both ends is not a random process at metaphase I and metaphase II (X2(specimen 1, meta I)=19.5; X2(specimen 1, meta II)=23.5; X2(specimen 2, meta I)=7.71; X2(specimen 2, meta II)=7.11; X2(specimen 3, meta I)=13.16; X2(specimen 3, meta II)=13.53 > CV(L=1; α=0.05)=3.84), and ii) the ends that are active during the first meiotic division become inactive during the second one (X2(specimen 1)=3.49; X2(specimen 2)=8.82x10-4; X2(specimen 3)=2.38 < CV(L=1;α=0.05)=3.84).
Photos (left) and diagrams (right) illustrating two alternative orientations of the autosomal pair with the NOR: a–b at metaphase I and c–d metaphase II in Microtomus lunifer. Chromosomes: grey; rDNA clusters: white.
So far, the cytogenetic analysis of 153 species from Reduviidae
reveals a chromosome diploid number that varies from 10 to 34, with
both simple and multiple sex chromosome systems (XY/XX, X0/XX, and
XnY/XnXn) (
It deserves attention that, even though Microtomus lunifer possesses the same diploid autosomal number as Microtomus conspicillaris,
both species differ in their sex chromosome system; the former has a
multiple sex chromosome system X1X2Y (male) whereas the latter presents a
simple sex chromosome system XY (male). The most common sex chromosome
system in Hemiptera
is the simple system XY/XX (male/female). Nevertheless, the other
simple system X0/XX, multiple systems (Xn0, XnY, XYn, XnYn) and
neo-systems are also reported (
It is generally accepted that multiple systems in Hemiptera are the result of fragmentation(s) of the X and/or Y chromosome(s) of an ancestral simple sex chromosome system (
In Hemiptera, autosomal bivalents are chiasmatic (except in a few families, such as Nabidae, Miridae, Cimicidae; see
Frequencies of cells at metaphase I and metaphase II showing the kinetic activity restricted to the NOR or not NOR ends of the largest autosomal pair.
| Specimen | Frequency | Metaphase I | Metaphase II | ||
|---|---|---|---|---|---|
| configuration a | configuration b | configuration c | configuration d | ||
| 1 | F1* | 24 | 66 | 40 | 7 |
| f1* | 0.27 | 0.73 | 0.85 | 0.15 | |
| 2 | F2* | 12 | 30 | 26 | 10 |
| f2* | 0.29 | 0.71 | 0.72 | 0.28 | |
| 3 | F3* | 70 | 120 | 51 | 20 |
| f3* | 0.37 | 0.63 | 0.72 | 0.28 | |
The basal position of Hammacerinae was earlier proposed by
A particular feature of both Microtomus
species is the presence of a minute chromosome pair with a different
meiotic behaviour from that of autosomes and sex chromosomes, the
so-called m-chromosomes. Most reports on the behaviour of the
m-chromosomes described them as asynaptic and achiasmatic throughout
early meiotic prophase after conventional staining squashed
spermatocytes. At diakinesis they approach each other, and at metaphase
I they are always associated end-to-end (touch-and-go pairing) forming a
pseudo-bivalent that segregates reductionally at anaphase I. However,
minor modifications of this typical male meiotic behaviour are found
among different taxa, particularly with regard to the size, the
pycnotic cycle, the meiotic behaviour, and the arrangement at both
metaphases I and II (
From diplotene onwards, the smallest chromosome pair of Microtomus lunifer appeared not only as structures resembling true bivalents (Figs 2d, e; 4b) (
Taking into account the meiotic behaviour of the m-chromosomes in Coreus marginatus and the presence of m-chromosome pair in Microtomus conspicillaris our results allow us to suggest that the minute chromosome pair of Microtomus lunifer could be considered a pair of m-chromosomes.
Up to now, Microtomus conspicillaris and Microtomus lunifer are the only two species within Reduviidae that possess a pair of m-chromosomes; thus, the presence of this pair could be a synapomorphy for the species of MicrotomusIlliger, 1807.
C- and fluorescent bandingsIn Hemiptera
early reports on C-positive heterochromatin showed that C-bands are
terminally located in some or all the chromosomes. However,
interstitial C-positive bands are described in a few species and some of
them correspond to secondary constrictions and NORs (
The use of fluorescent DNA-binding dyes with
different specificities allows a better characterization of
heterochromatic regions in terms of their relative enrichment with AT or
GC base pairs. Most reports referring to heterochromatin
characterization on hemipteran species describe C-bands as DAPI bright
and CMA3 dull. The presence of a CMA3 bright band was detected in a few
species at interstitial or terminal position, either on autosomes or
sex chromosomes, and they are generally associated to NORs (
In Reduviidae the location of NORs was analysed in only 14 species belonging to the subfamilies Harpactorinae (2 species) and Triatominae (12 species) by Ag-NOR, fluorescent banding and/or FISH with rDNA probes (18S, 26S or 45S). The present paper brings the first information about the number and chromosomal location of ribosomal gene clusters in Hammacerinae. Using rDNA-FISH we show here that Microtomus lunifer has an rDNA cluster, which is located at one terminal region of the largest autosomal pair.
In Microtomus luniferthe
NOR is associated with a small CMA3 bright band. The results of the
fluorescent banding and FISH in this species agree with those described
for Rhodnius pallescens Barber, 1932 (
Taking into account the data on the number and location of rDNA clusters along with the type of sex chromosome systems in Reduviidae,
we can observe different patterns of rDNA distribution. The NOR is
generally located at terminal position on the X chromosome, or on both X
and Y chromosomes in the species that have XY sex chromosome system. On
the other hand, in most cases the NOR is placed at terminal position
on an autosomal pair in the species with multiple sex chromosome systems
(XnY). Providing that the ancestral male karyotype of Reduviidae
had 2n=30=28+XY, the NOR would have been at a terminal region of the
sex chromosomes. Thus, a single pair of NOR-autosomes in species with
multiple sex systems (XnY) might be due to the ability of NOR to change
its number and position (
In two species of Coreidae, Carlisis wahlbergi Stål, 1858and Camptischium clavipes(Fabricius, 1803), most crossovers occurred in the distal half of the NOR-bivalent (
In Microtomus luniferthe location of the NOR at one chromosome end was used as a chromosome marker that allowed us to discern both ends, determine whether both terminal regions could be kinetically active and analyse the behaviour of autosomes during both meiotic divisions. The hypotheses which were tested are the followings: i) the kinetic activity of both ends at both meiotic divisions is a random process, and ii) those regions that were active during anaphase I become inactive during anaphase II and vice versa. From our results it can be concluded that both terminal regions are able to develop kinetic activity at first and second meiotic divisions, but the election of the kinetic end is not a random process. In addition, those chromosome ends that show kinetic activity in the first meiotic division are inactive in the second one, and vice versa.
The identification of the factor/s and the
mechanism/s involved in the restriction of the kinetic activity to only
one chromosome/chromatid end in holokinetic chromosomes of Hemiptera remains unsolved. The presented results here together with previous papers (
In summary, the analysis of meiosis, the determination of the distribution, number and location of heterochromatin blocks and rDNA loci could be useful for the taxonomic identification of species, the analysis of karyotype evolution, and for a better knowledge of chromosome structure and organization.
This work was funded by grants UBACyT X164 of University of Buenos Aires, PIP 0281 of Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), and PICT 2007-00635 of ANPCyT from Argentina. MG Poggio, MJ Bressa, and AG Papeschi thank CONICET. We wish to thank Dr. MC Melo and Lic. YM Provecho for taxonomic identification of the specimens included in the study, and for collecting specimens, respectively.