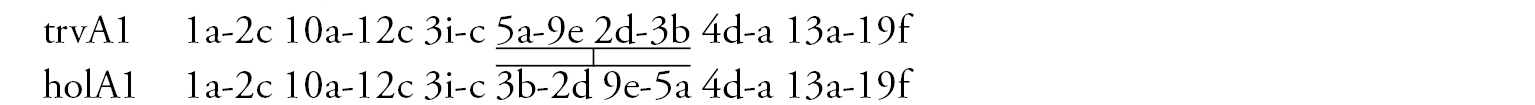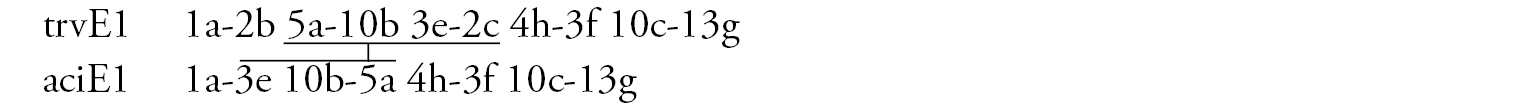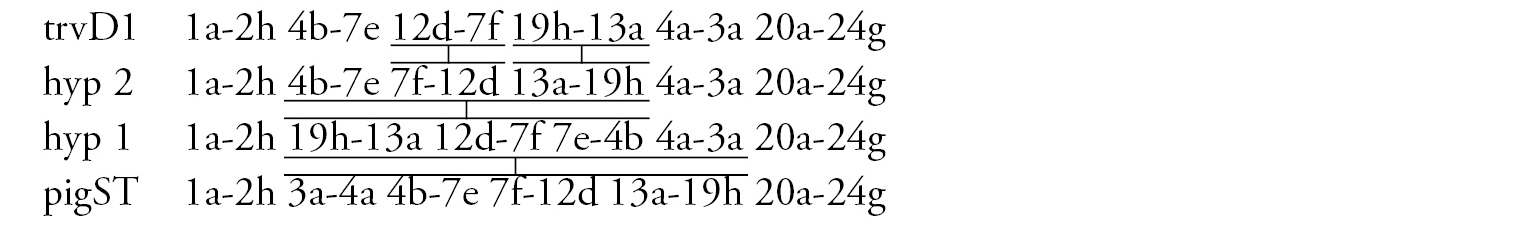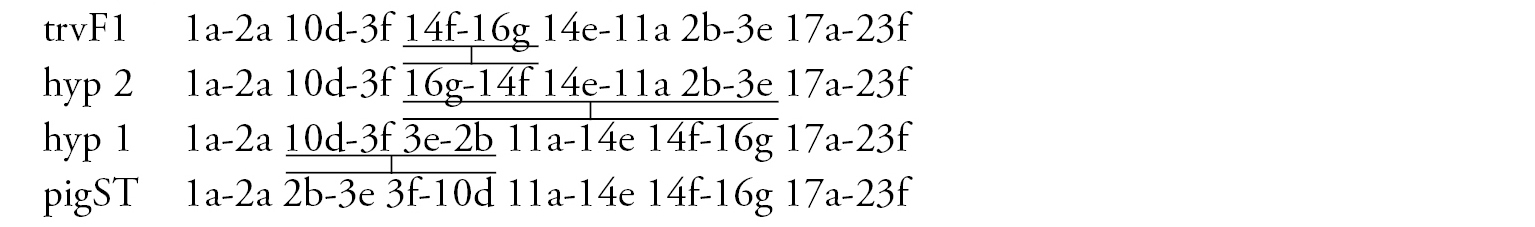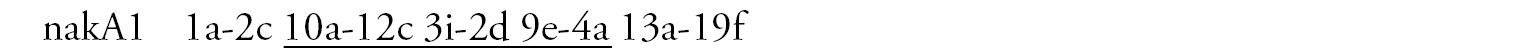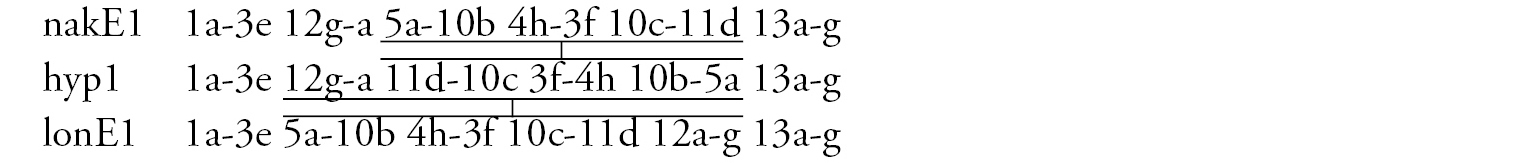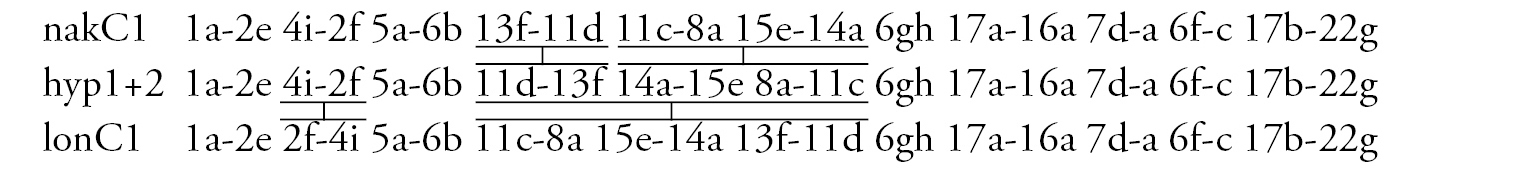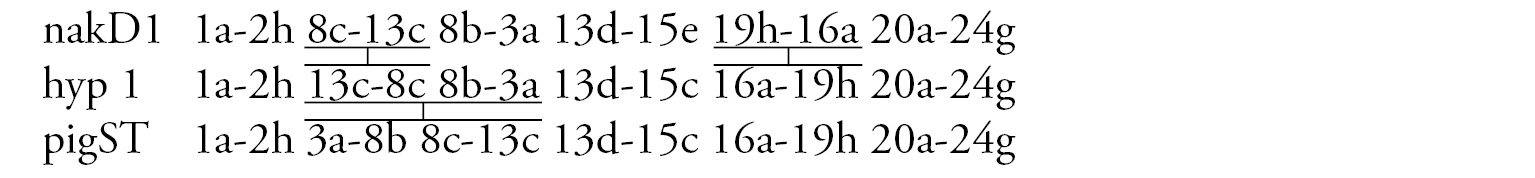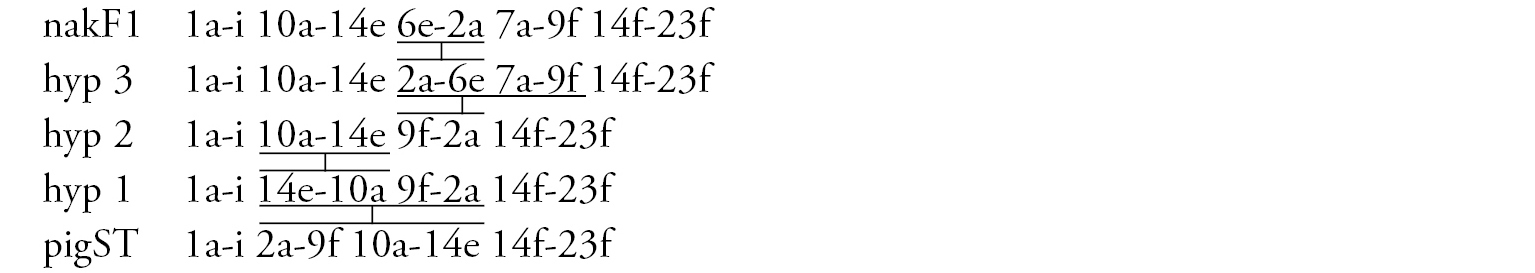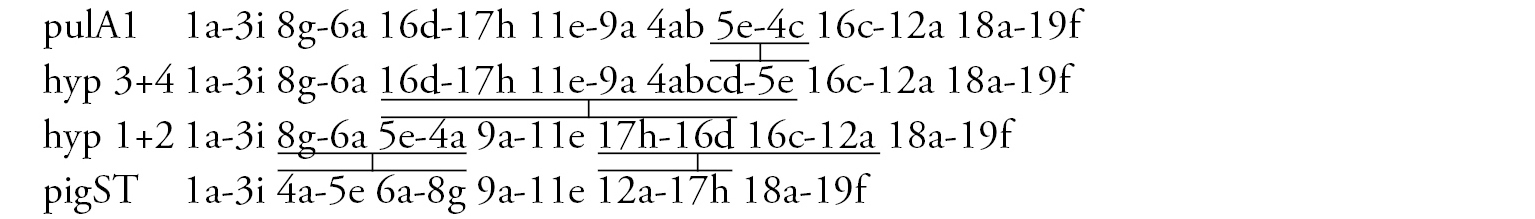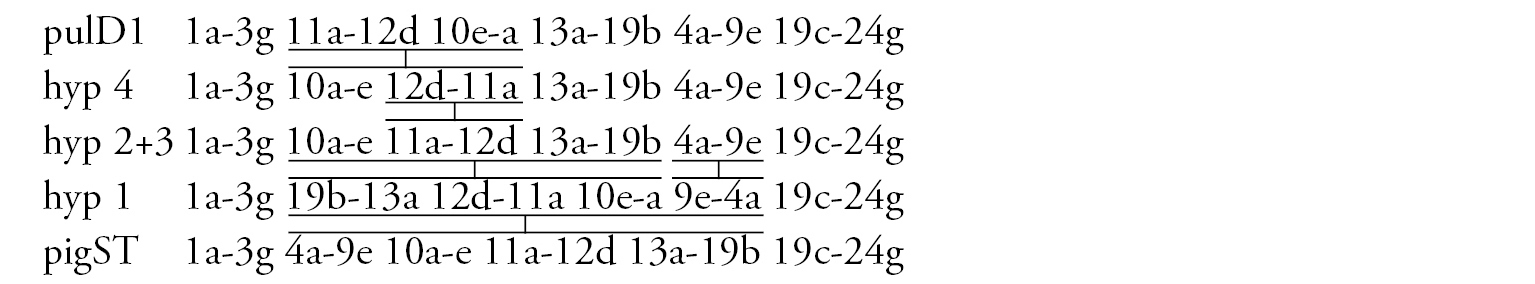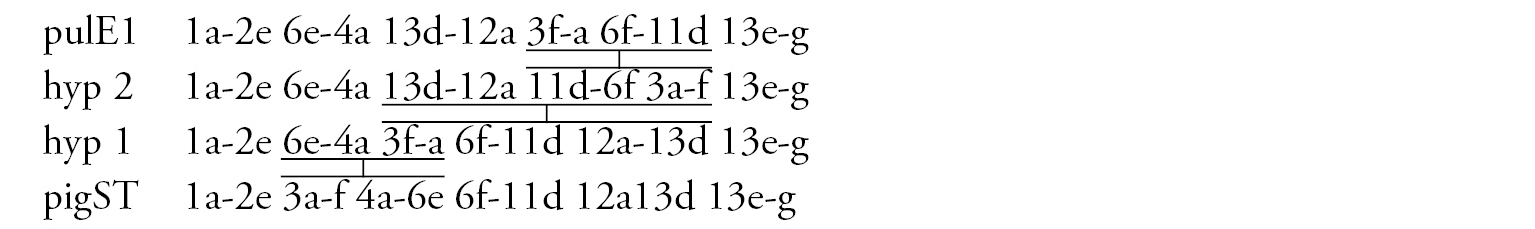(C) 2011 Wolfgang F. Wülker. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
The karyotypes of six African Chironomus species (Chironomus alluaudi Kieffer, 1913, Chironomus transvaalensis Kieffer, 1923, Chironomus sp. Nakuru, Chironomus formosipennis Kieffer, 1908, Chironomus prope pulcher Wiedemann, 1830, Chironomus sp. Kisumu) were investigated; four of these karyotypes were described for the first time (Chironomus sp. Nakuru, Chironomus formosipennis, Chironomus prope pulcher, Chironomus sp. Kisumu). Of the six Chironomus karyotypes, three had “pseudothummi” cytocomplex chromosome arms combinations AE CD BF G (Chironomus alluaudi, Chironomus transvaalensis, Chironomus sp. Nakuru), two had “thummi”cytocomplex arms combinations AB CD EF G (Chironomus formosipennis, Chironomus prope pulcher), and one had “parathummi”armcombinations AC BF DE G (Chironomus sp. Kisumu). Thus, three of the ten main cytocomplexes known were detected in Africa. Detailed photomaps of all chromosome arms, with the exception of arms B and G, were prepared for the karyotypes of Chironomus alluaudi, Chironomus transvaalensis, Chironomus sp. Nakuru, Chironomus prope pulcher; the karyotypes of Chironomus formosipennis, Chironomus sp. Kisumucould only be fragmentarily mapped.
Endemic African banding sequences were characteristic
for most of the chromosomal arms in all species studied. However,
basic sequences, which can be present in different Chironomus species on different continents (Wülker, 1980;
"Chironomus, karyotype, banding sequences, chromosomal polymorphism, chromosomal evolution
As shown by cytogenetic analysis of chromosomal
evolution, the divergence of animal karyotypes during speciation was
mainly mediated by para- and pericentric inversions, altering the gene
orders in linkage groups (
By global analysis of banding sequences in Eurasia,
North and South America, Australia, we have traced banding sequence
changes during Chironomus species divergence and continent dispersal (
This paper contains full descriptions of the karyotypes of six African Chironomus species (Chironomus alluaudi, Chironomus transvaalensis, Chironomus sp. Nakuru, Chironomus formosipennis, Chironomus prope pulcher, Chironomus sp. Kisumu). Among them four karyotypes are described for the first time (Chironomus sp. Nakuru, Chironomus formosipennis, Chironomus prope pulcher, Chironomus sp. Kisumu). Detailed photomaps of arms A, C, D, E, and F are presented for Chironomus alluaudi, Chironomus transvaalensis, and Chironomus sp. Nakuru. The chromosome arms could be mapped only partly for Chironomus formosipennis, Chironomus prope pulcher and Chironomus sp. Kisumu.
The presence of further basic banding sequences in the karyotypes of African Chironomus species was discovered, along with endemic continent-specific (Ethiopian region) sequences.
Evolutionary divergence of “thummi“ and “pseudothummi” cytocomplexes is discussed.
Material and methodsForth instar larvae of African Chironomus species were used for karyotype study. 35 years ago, one of us (W.W.) had the opportunity to visit Kenya (22.12.1975–16.01.1976). From a base at the house of relatives in Nairobi, he went with family (wife and 3 sons) to collect chironomids to the west to Lake Nakuru and Lake Victoria, to the north to Abrader Mount Ca. 3000 m above N.N., and to the southeast to Tsavo National park, Mombasa and vicinity. Other material was contributed by colleagues: Mount Elgon and Lake Naivasha (Peter N. Cox), Mount Kenya, near 4350 m (scientific excursion of University Erlangen, Germany, under Prof. Dr. Rüppell), Zigi River, Tanzania (Dr. J. Grunewald). The list of collection sites of Chironomus larvae is presented in Table 1. We have not identified species Chironomus sp. Nakuru and Chironomus sp. Kisumu, but the study of the banding sequences of their karyotypes was very important for purpose of our paper.
Collection sites and number of specimens of African Chironomus species.
| Species | Collection sites | Collection date | Collector | Number of specimens |
|---|---|---|---|---|
| Chironomus alluaudi | Kenya: drinking troughs brooks and pools at Endebess/Mt. Elgon, mountain lakes W of Nakuru, Aberdare mountains up to 3300 m, ponds at MtKenya 4350 m, near Limuru (north of Nairobi), Athi-river south of Nairobi, Amboseli-park | 29.12.75 03.01.76 12.01.76 13.01.76 10.01.76 | P. N. Cox, W.Wülker. G. Rüppell W. d’Oleire-Oltmanns, H.Koehler W. Wülker | 120 |
| Chironomus transvaalensis | Kenya: pool east Lake Victoria, north and south Athi-river near Nairobi, west of Mombasa, Tansania: Kikuwi-river | 27.12.75 13.01.76 | W. Wülker; J. Grunewald | 115 |
| Chironomus sp. Nakuru | Kenya: brook south east Lake Nakuru | 26.12.75 | W. Wülker | 9 |
| Chironomus formosipennis | Kenya: Lake Naivasha, Tansania: Zigi-river, running waters | P. N. Cox, J. Grunewald | 15 | |
| Chironomus prope pulcher | Kenya: two pools in short distance, River Athi south of Nairobi | 13.01.76 | W. Wülker | 6 |
| Chironomus sp. Kisumu | Kenya: flat pools 10 and 22 km east Lake Victoria, together with Chironomus transvaalensis, brook in Amboseli-park (Kilaguni) | 27.12.75 10.01.76 | W. Wülker | 7 |
Larvae were fixed in ethanol-glacial acetic acid (3:1). The technique of chromosome preparation was as usual (
To trace the relationship of African Chironomus banding sequences with sequences from other continents, we compared them with known basic sequences; if basic sequences for some of species were unknown, we compared them with Chironomus piger standard (ST).
We have pointed to previous literature on morphological characteristics of species studied at the beginning of each species description. Most part of the material (larvae, pupa, adults and karyotype slides) is now deposited in Zoologische Staatssammlungen in Münich (Germany).
Equipment of the Center of Microscopy Analysis of Biological Objects of SB RAS in the Institute of Cytology and Genetics (Novosibirsk) was used in accomplishment of this work: microscope “Axiokop” 2 Plus, CCD camera AxioCam HRc, software package AxioVision 4 (Zeiss, Germany).
Results
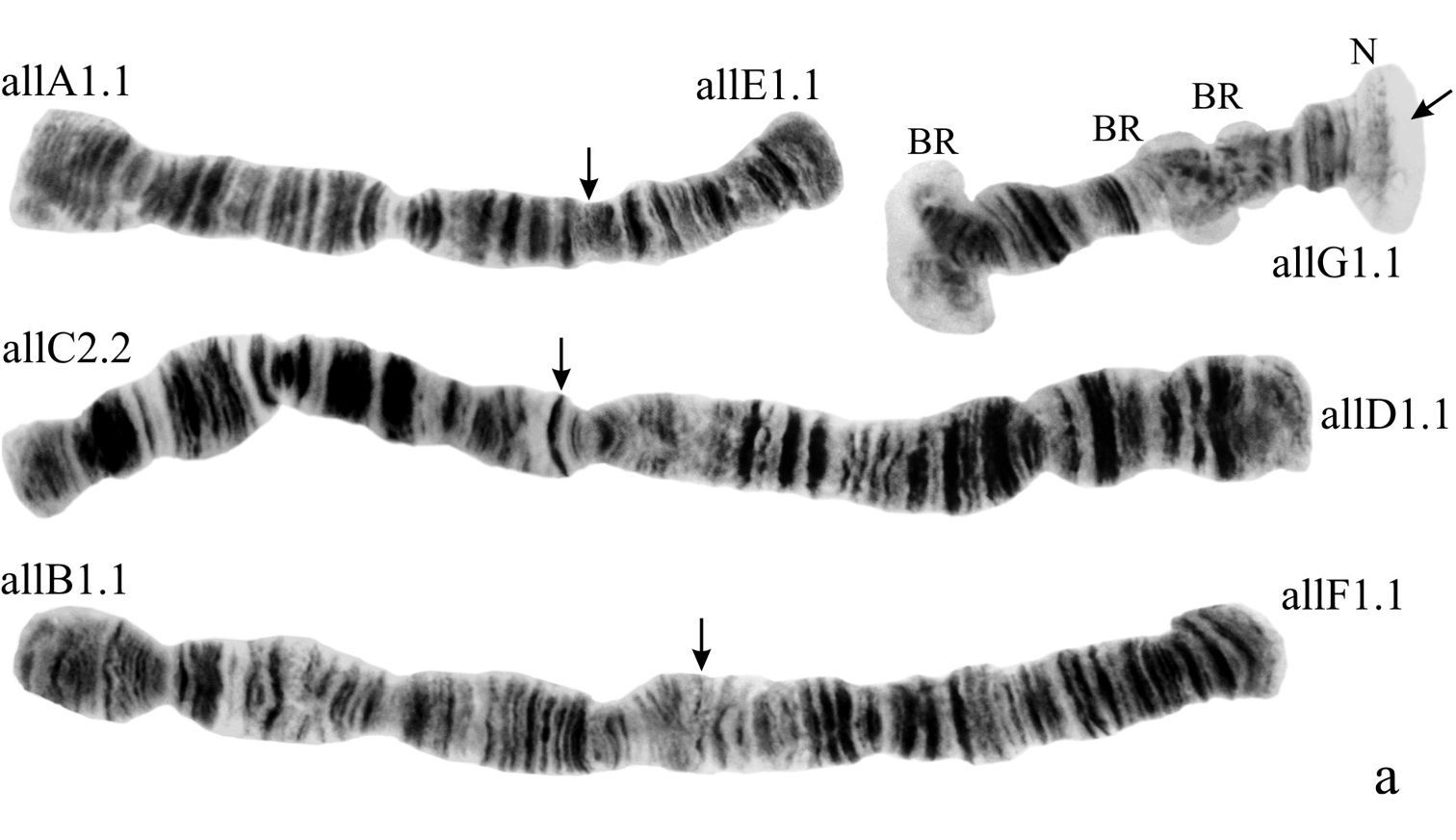
(Fig. 1a). Haploid number n=4, arm combination AE CD BF G (“pseudothummi” cytocomplex), centromere bands not heterochromatinized, nucleolus in arm G (terminal), at least 3 Balbiani Rings (BRs) on arm G, inversion polymorphism in arms C and G.
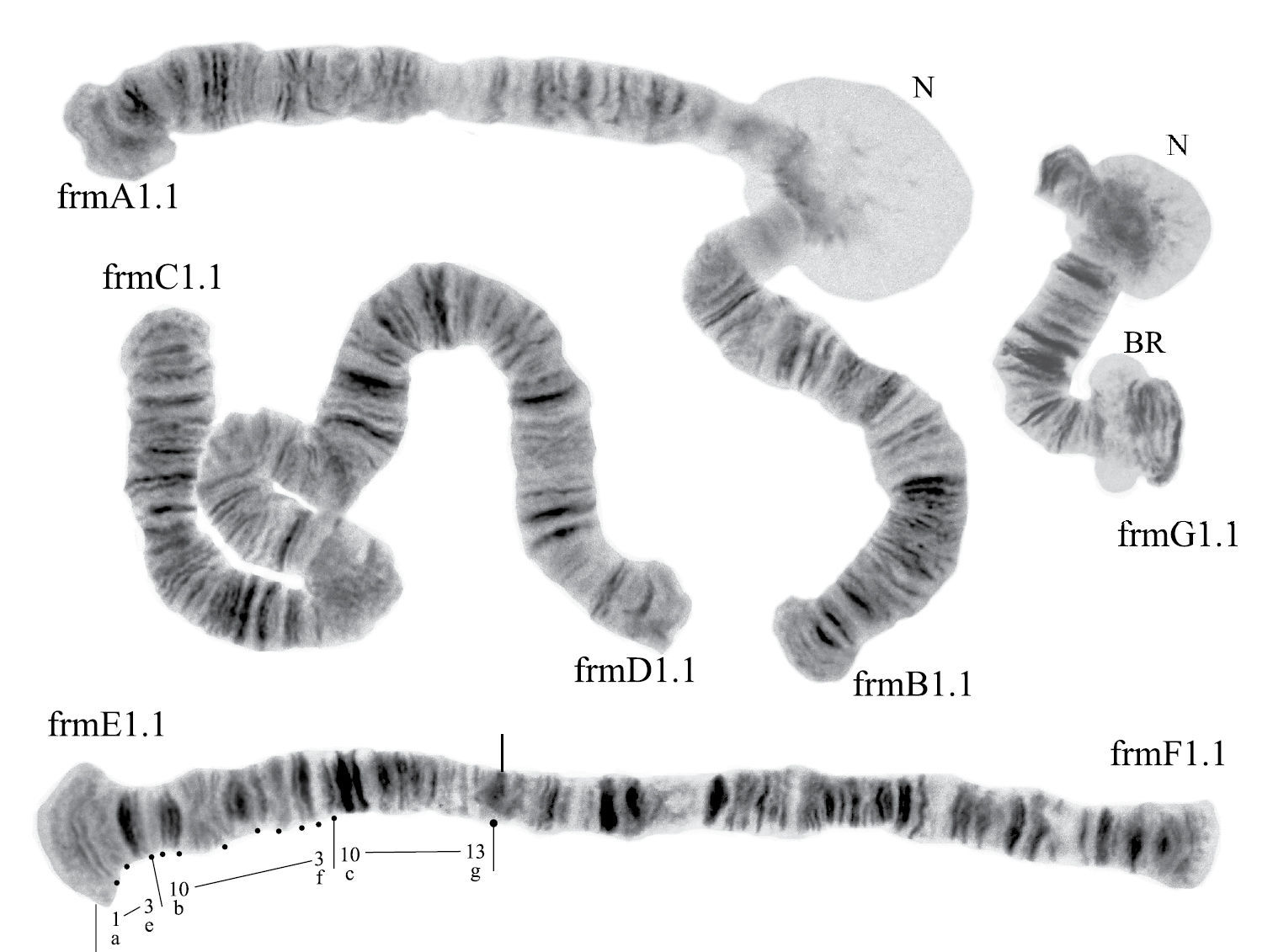
Karyotype of Chironomus alluaudi. In this and all other Figures: allA1.1, allE1.1 etc. – symbols of arm and homozygous genotypic combinations N – nucleolus BR – Balbiani ring, arrows show centromeric bands, brackets near chromosome arms show inversions.
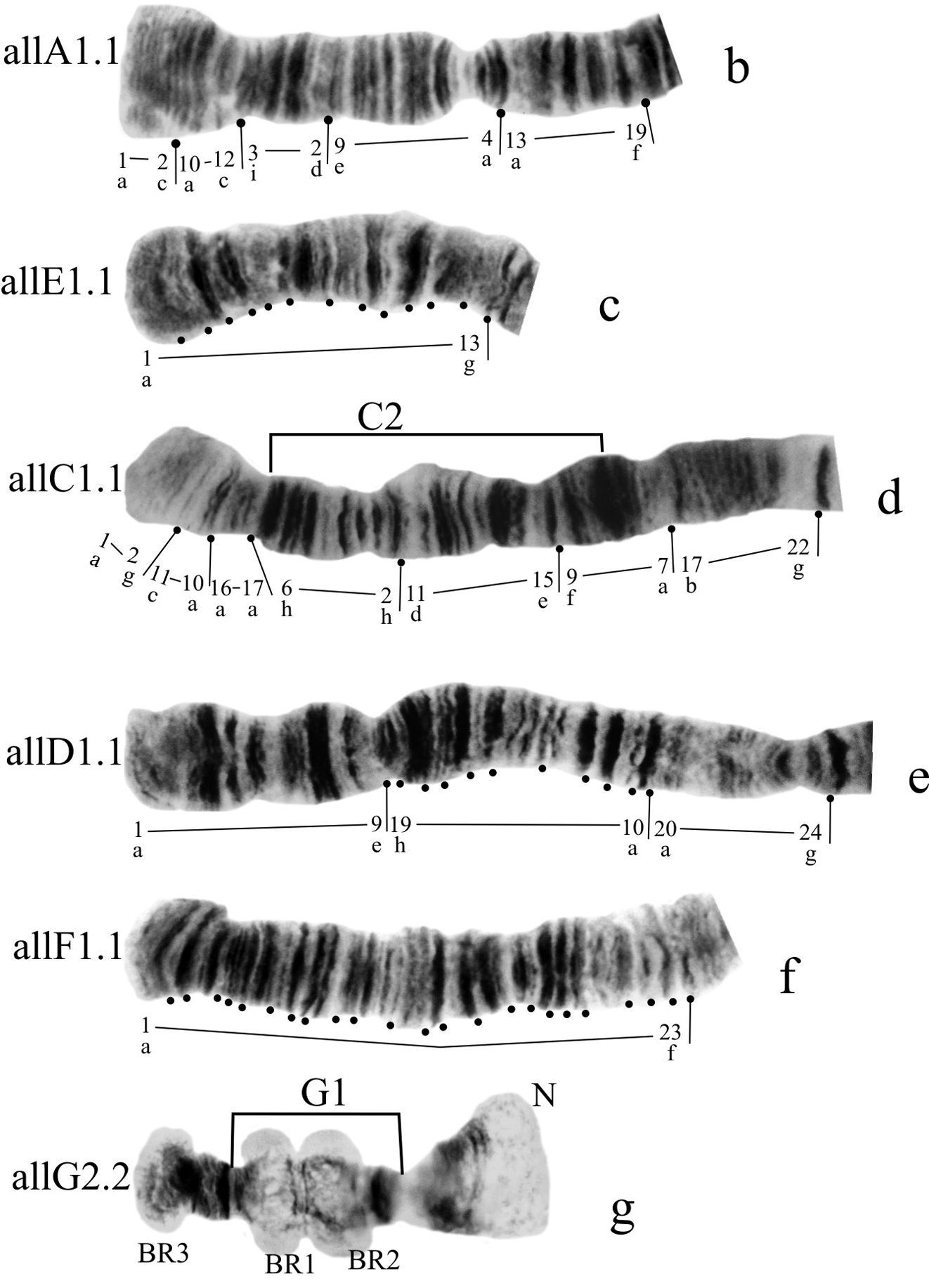
Banding sequences (Fig. 1b-g)
Homozygous banding sequences of Chironomus alluaudi in arms A, E, C, D, F and G. The designations are the same as in Fig. 1.
Arm A (Fig. 1b) has the sequence all A1 identical with the main sequence of arm A found in many Chironomus species (Chironomus holomelas Keyl, 1961, Chironomus melanescens Keyl, 1961, etc.) and it is considered a cosmopolitan basic sequence (holA1).
Arm E (Fig. 1c, 7b) has the sequence allE1 identical with Chironomus piger ST (cosmopolitan basic sequence).
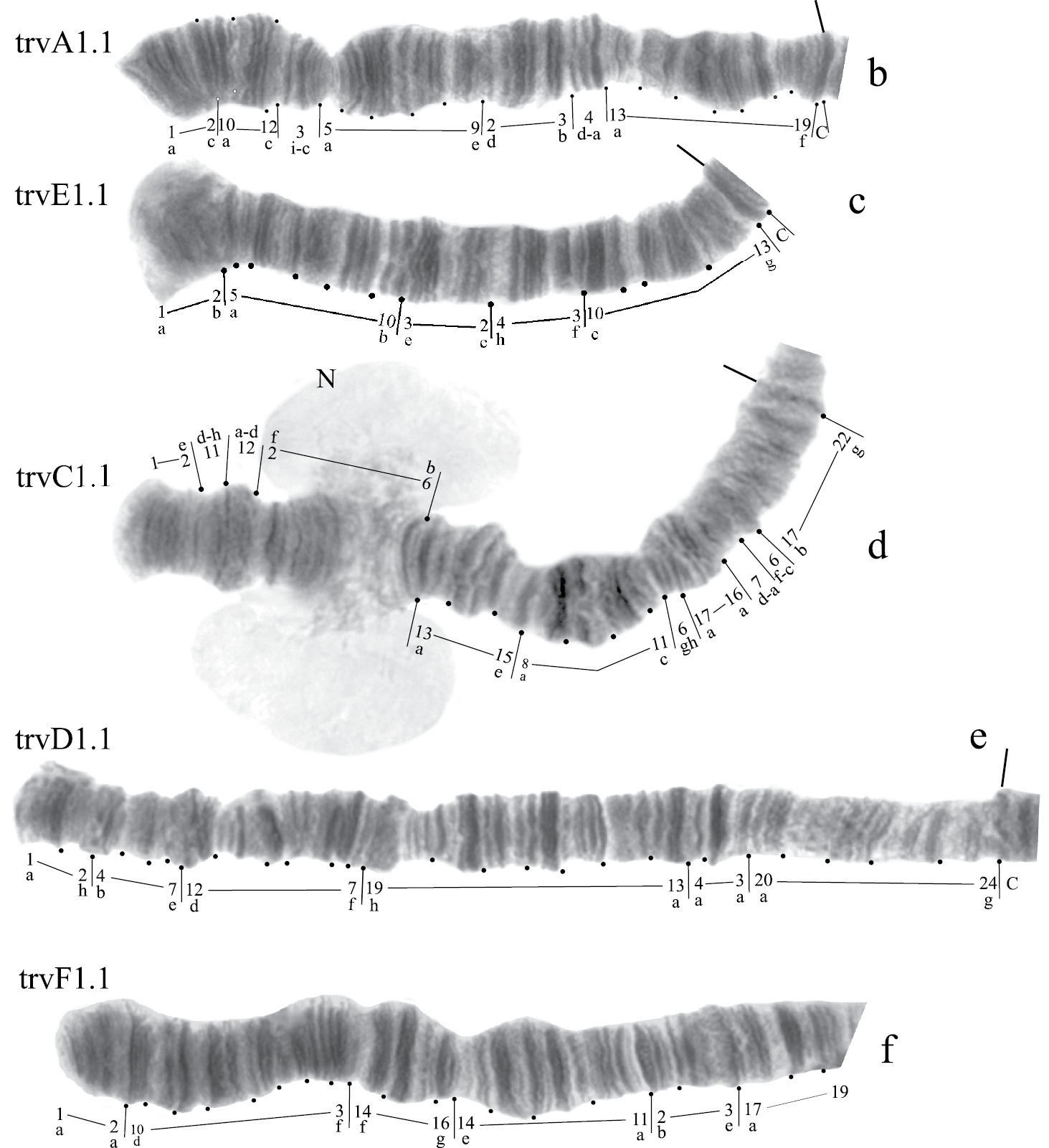
Karyotype of Chironomus transvaalensis. p – puff and the designations are the same as in Fig. 1
Arm C (Fig. 1d) has two sequences, allC1 and allC2, differing by a simple inversion. The sequence allC1 differs greatly from the basic sequence in arm C; therefore we have compared it with Chironomus piger ST: differing by seven inversion steps from pigST:
Arm D (Fig. 1e) has single sequence allD1 differing by one inversion step from pigST:
Arm B (Fig. 1a) not mapped, monomorphic. The common BR is not developed.
Arm F (Fig. 1f) has the sequence allF1, identical with pigST (cosmopolitan basic sequence).
Arm G (Figs 1a, g) not mapped, has two sequences allG1 and allG2 differing by one simple inversion in the central part of arm G, including two of the Balbiani rings.
In total, the banding sequence pool of Chironomus alluaudi contains 9 sequences. Six of them endemic for Africa (Ethiopian sequences), three of them (allA1, allE1, allF1) belong to the category of cosmopolitan basic sequences. Chironomus alluaudi can be considered asa Chironomus species with a primitive karyotype (Wülker, 1980, 2010).
“thummi-type” (no tubuli laterales) on abdominal segment VII). Mentum with high lateral tooth, median tooth as in other Chironomus species, pectin epipharyngis about 11 teeth, antenna black with 4 segments, paralabial plates about 40 striae.
different places in Africa (
Mc Lachlan 1969, 1971: larva and pupa.
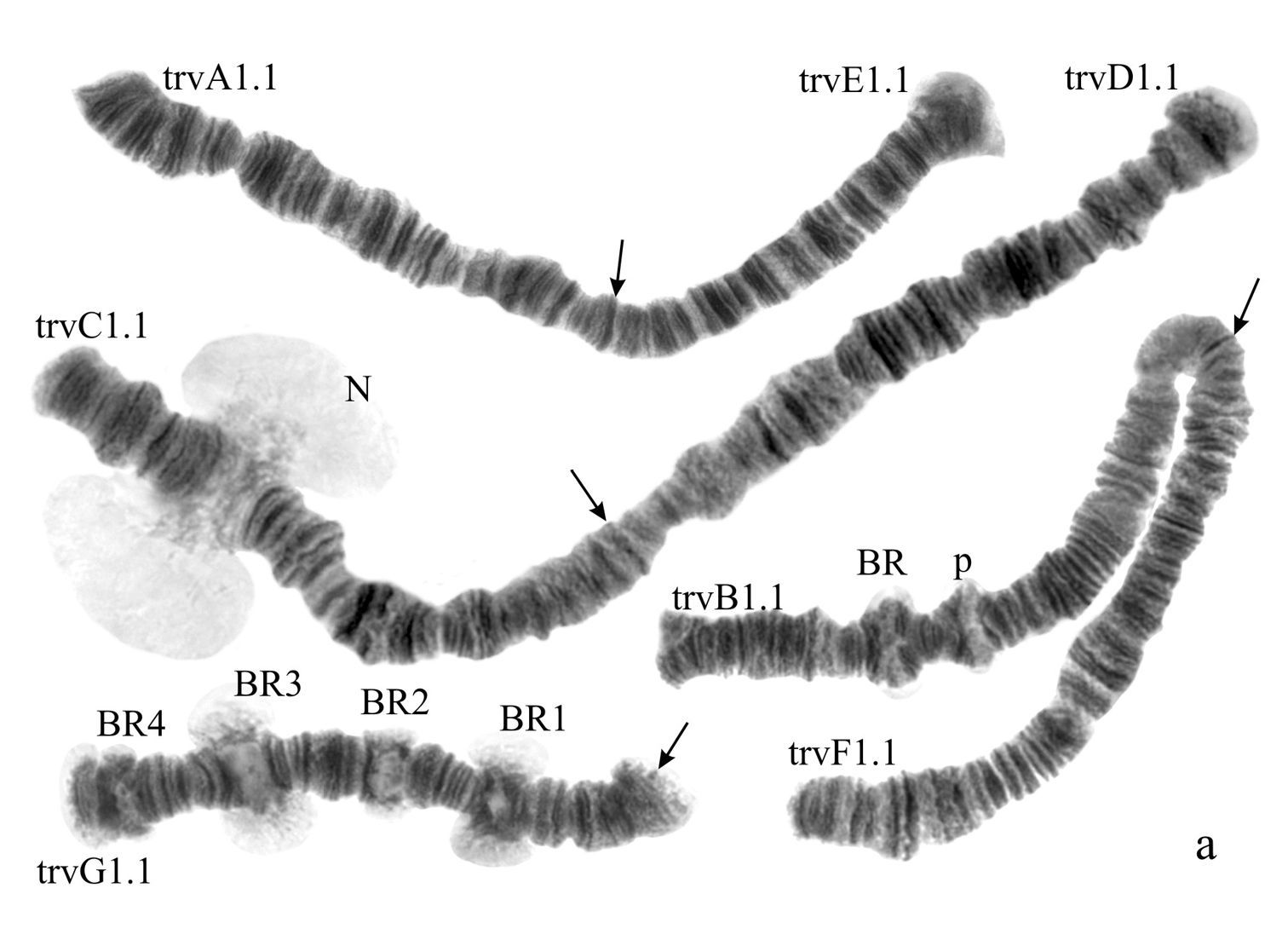
(Fig. 2a). Haploid number n=4, arm combination AE CD BF G (“pseudothummi” cytocomplex), centromeric bands not heterochromatinized, nucleolus in arm C, inversion polymorphism in arms C and G.
Banding sequences (Fig. 2b-f).
Homozygous banding sequences of Chironomus transvaalensis in arms A, E, C, D and F.
Arm A (Fig. 2b) has the sequence trvA1, differing by only one inversion step from the basic sequence holA1.
Arm E (Fig. 2c) has the banding sequence trvE1, differing only by one step from basic sequence aciE1 (Chironomus acidophilus Keyl, 1960 etc.)
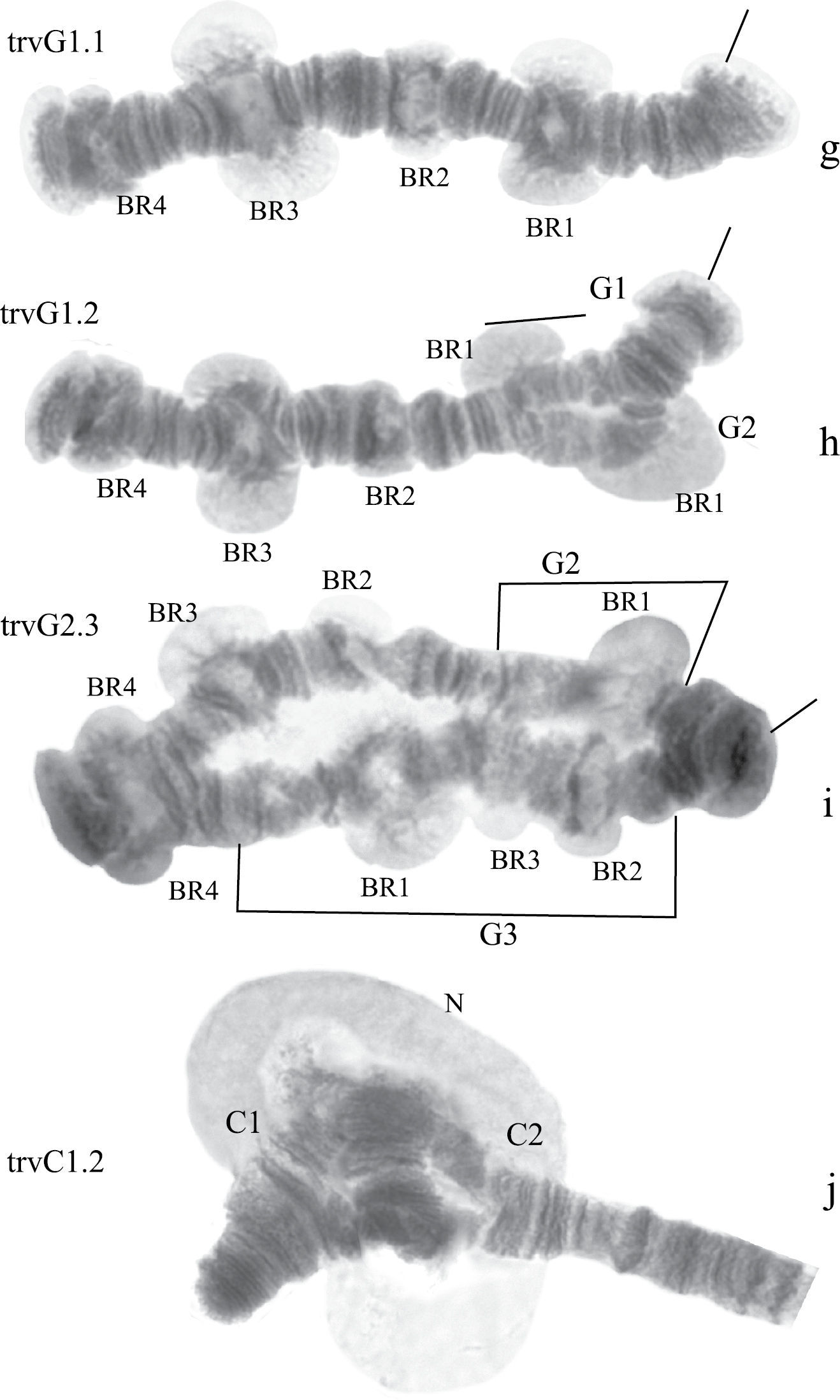
Arm C (Fig. 2d, j) has two banding sequences, trvC1 and trvC2, differing by one simple inversion (Fig. 2j). The sequence trvC1 is formed by four inversion steps from a basic sequence, (lonC1), found in several Chironomus species (Chironomus longistylus Goetghebuer, 1921, Chironomus anthracinus Zetterstedt, 1860 etc.).
Homozygous and heterozygous banding sequences of Chironomus transvaalensis in arm G (g–i) and heterozygous inversion in arm C (j). Brackets above arms indicate the localization of inversions. The designations are the same as in Fig. 1.
Arm D (Fig. 2e) has the sequence trvD1 differing from pigST by four inversion steps.
Arm B (Fig. 2a) not mapped, monomorphic. BR is well developed.
Arm F (Fig. 2f) has the banding sequence trvF1 differing from cosmopolitan basic pigST by three inversion steps.
Arm G (Fig. 2g-i) has three banding sequences, trvG1, trvG2, and trvG3. The sequence trvG2 differs from trvG1 by a short inversion in the region BR1 (Fig. 2h); the sequence trvG3 – by long inversion of central part of arm G (Fig. 2i). Both last sequences were found as heterozygotes. There are four Balbiani rings.
In total, the banding sequence pool of Chironomus transvaalensis contains 10 sequences, all of them are Ethiopian endemic sequences.
tubuli laterales at abdominal segment VII. Other characters - Mc Lachlan, 1969.
various places in Africa,
Wülker, 1980, banding pattern of arms A, E, and F. This species was not identified as well as Chironomus sp. Kisumu because there was no additional possibility to collect larvae for rearing. However, the study of Chironomus sp. Nakuru karyotype was very important for comparative analysis of Ethiopian Chironomus banding sequences with Chironomus sequences of the other continents.
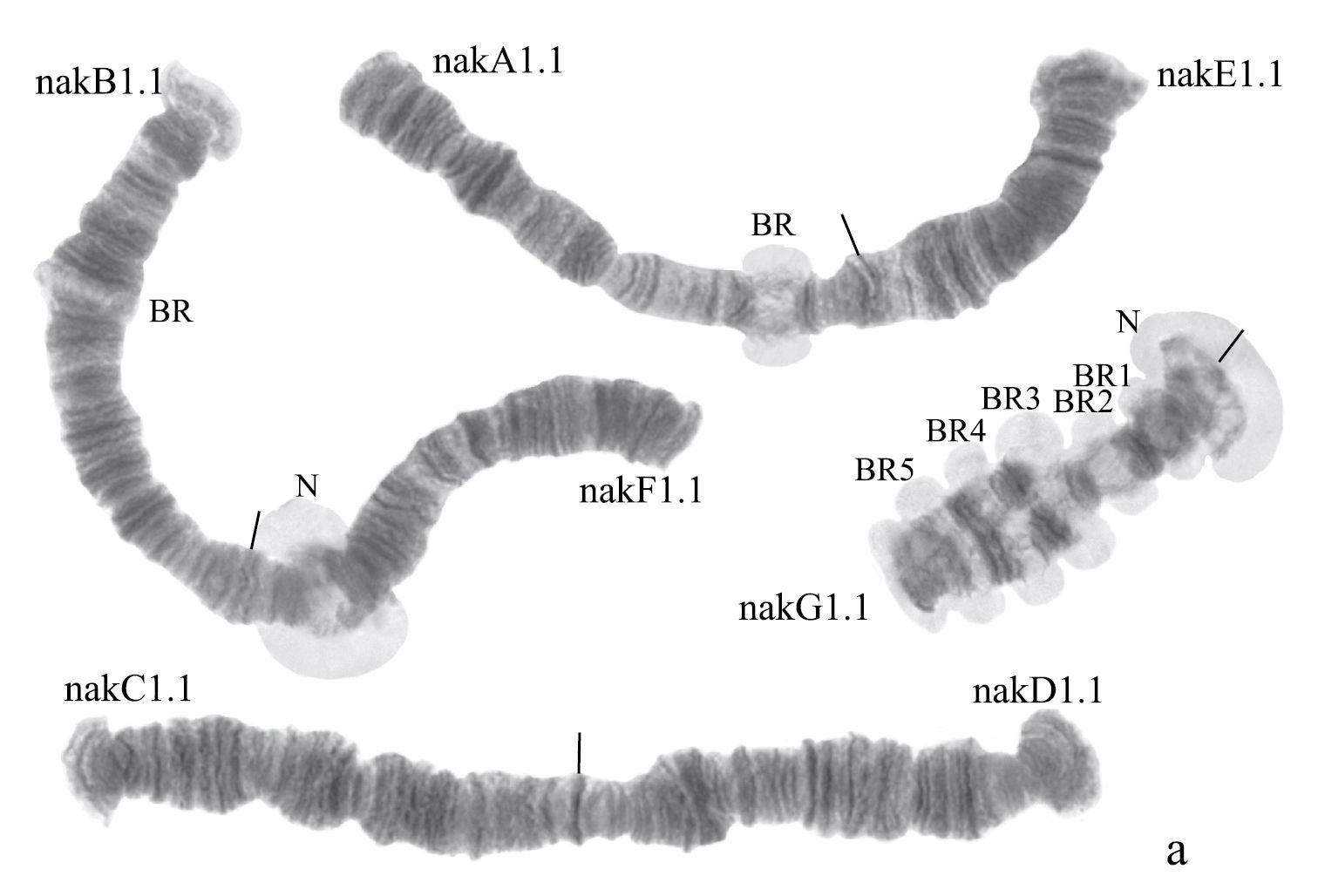
(Fig. 3a). Haploid number n=4, arm combination AE CD BF G (“pseudothummi” cytocomplex), centromeric bands not heterochromatinized, nucleoli on arms F and G, Balbiani rings on arms G, B, and A. Chromosomal polymorphism was not recorded.
Karyotype of Chironomus sp. Nakuru. The designations are the same as in Fig. 2a.
Homozygous banding sequences of Chironomus sp. Nakuru in arms A, E, F, C and D. The designations are the same as in Fig. 1.
Banding sequences (Fig 3b-f).
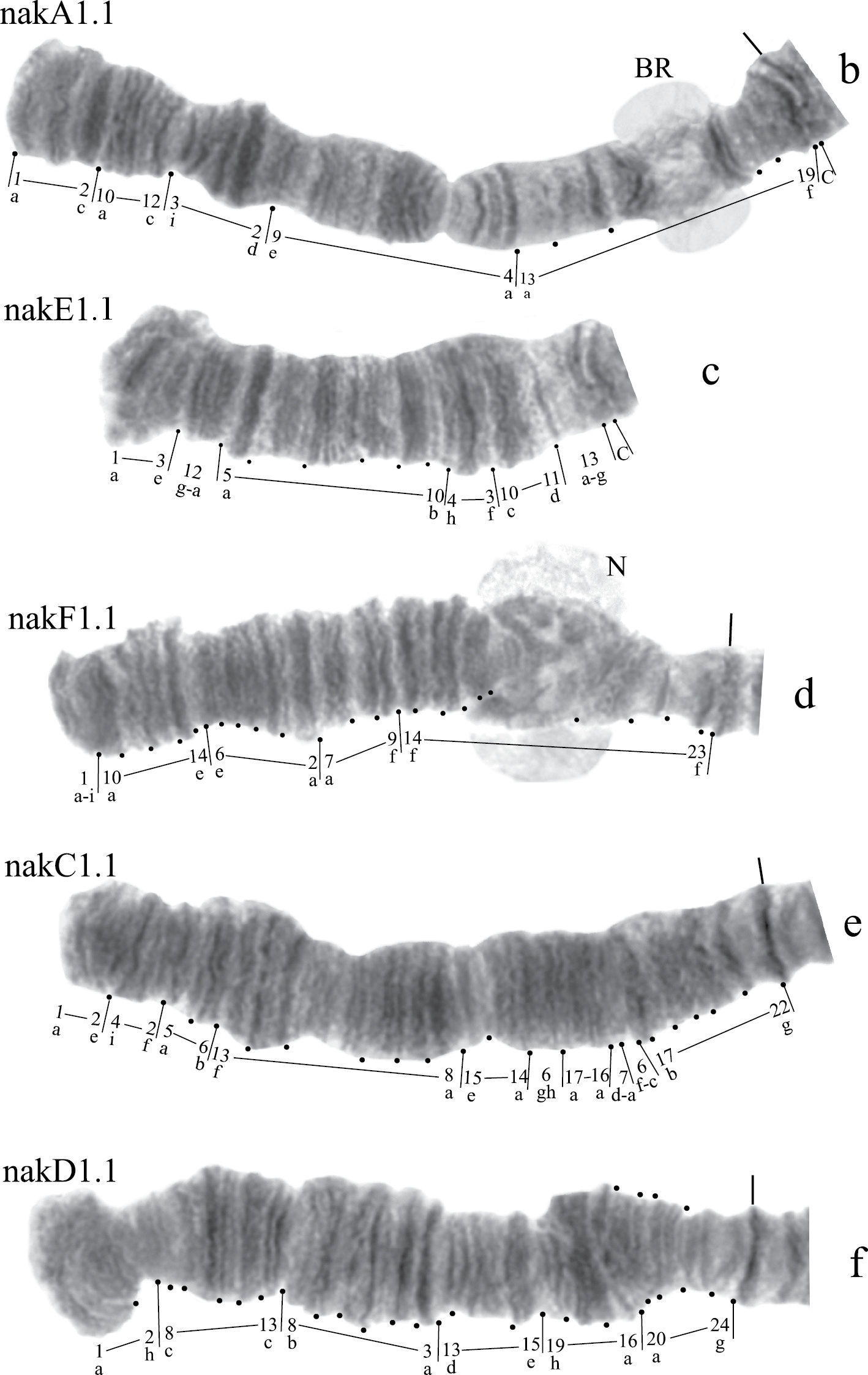
Arm A (Fig. 3b) has the banding sequence nakA1 identical with cosmopolitan basic sequence found in many species (Chironomus holomelas, Chironomus melanescens, etc.)
Arm E (Fig. 3c) has banding sequence nakE1 differing by two inversion steps from the cosmopolitan basic sequence lonE1 (Chironomus longistylus, Chironomus anthracinus etc.).
Arm C (Fig. 3e) has the sequence nakC1 differing by four inversion steps from basic pattern lonC1 (Chironomus longistylus, Chironomus anthracinus, etc.) and by seven inversion steps from Chironomus piger ST (Fig. 7b).
Karyotype of Chironomus formosipennis. The designations are the same as in Fig. 1.
Arm D (Fig. 3f) has the banding sequence nakD1 differing from pigST by three inversion steps.
Arm B (Fig. 3a) not mapped, monomorphic. The common BR is not developed.
Arm F (Fig. 3d) has the banding sequence nakF1 formed by four inversion steps from pigST.
The arm F of Chironomus sp. Nakuru has a nucleolus in region 17–19.
Arm G (Fig. 3a) has the banding sequence nakG1. It differs from the most of Chironomus species arm G by numerous Balbiani rings. It is possible to suggest that some of them can be nucleoli. But it is often impossible to differentiate nucleoli and Balbiani rings without electron microscopy or in situ hybridization.
In total, seven banding sequences are found in sequence pool of Chironomus sp. Nakuru, six chromosomal arms have Ethiopian endemic sequences, and one arm (A) a cosmopolitan basic sequence.
long tubuli laterales at abdominal segment VII, extremely long antenna, gula light, no dark stripe on clypeus.
brook to SE of Lake Nakuru, Kenya
(Fig. 4). Haploid number n=4, arm combinations AB CD EF G (“thummi” cytocomplex), centromeric bands not heterochromatinized, nucleoli in arms A and G, Balbiani ring in arm G. Chromosomal polymorphism was not recorded.
Banding sequence was determined only in arm E. The sequence frmE1 was identical with the cosmopolitan basic pattern, aprE1 (as in Chironomus aprilinus Meigen, 1818)
long tubuli laterales at abdominal segment VII. Other characters -
Lake Naivasha, Kenya, Zigi-river, Tanzania, running waters.
The association to this species is based on one male adult from the collecting sites of the larvae.
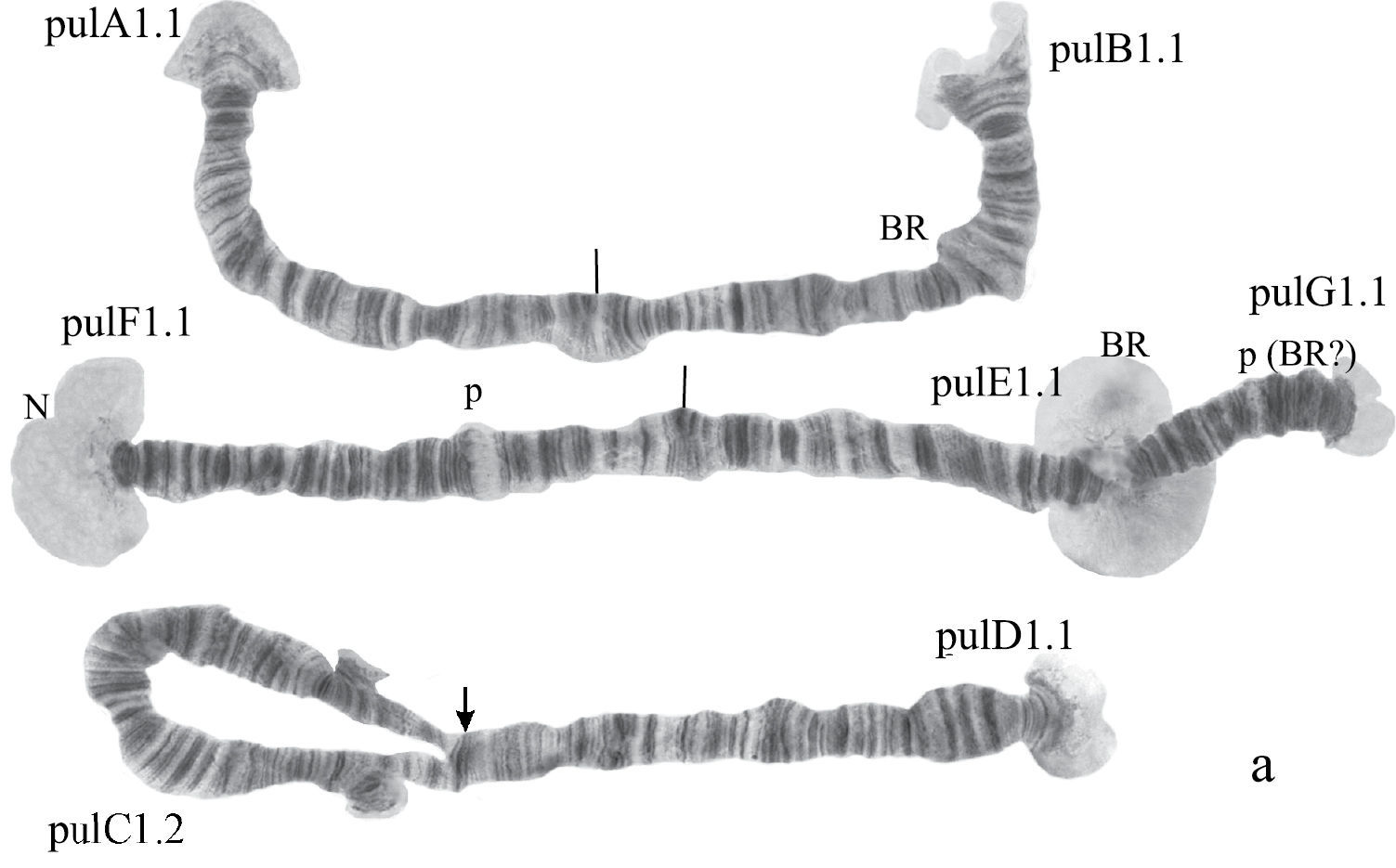
(Fig. 5a). Haploid number n=3, arm combination AB CD FEG (modified “thummi” cytocomplex), centromeric bands not heterochromatinized, nucleolus in arm F (at the very telomeric end) and nucleolus-like bodies at the ends of arms A, B, E; Balbiani rings are in arms G and B. Chromosomal polymorphism in arm C (Fig. 5a).
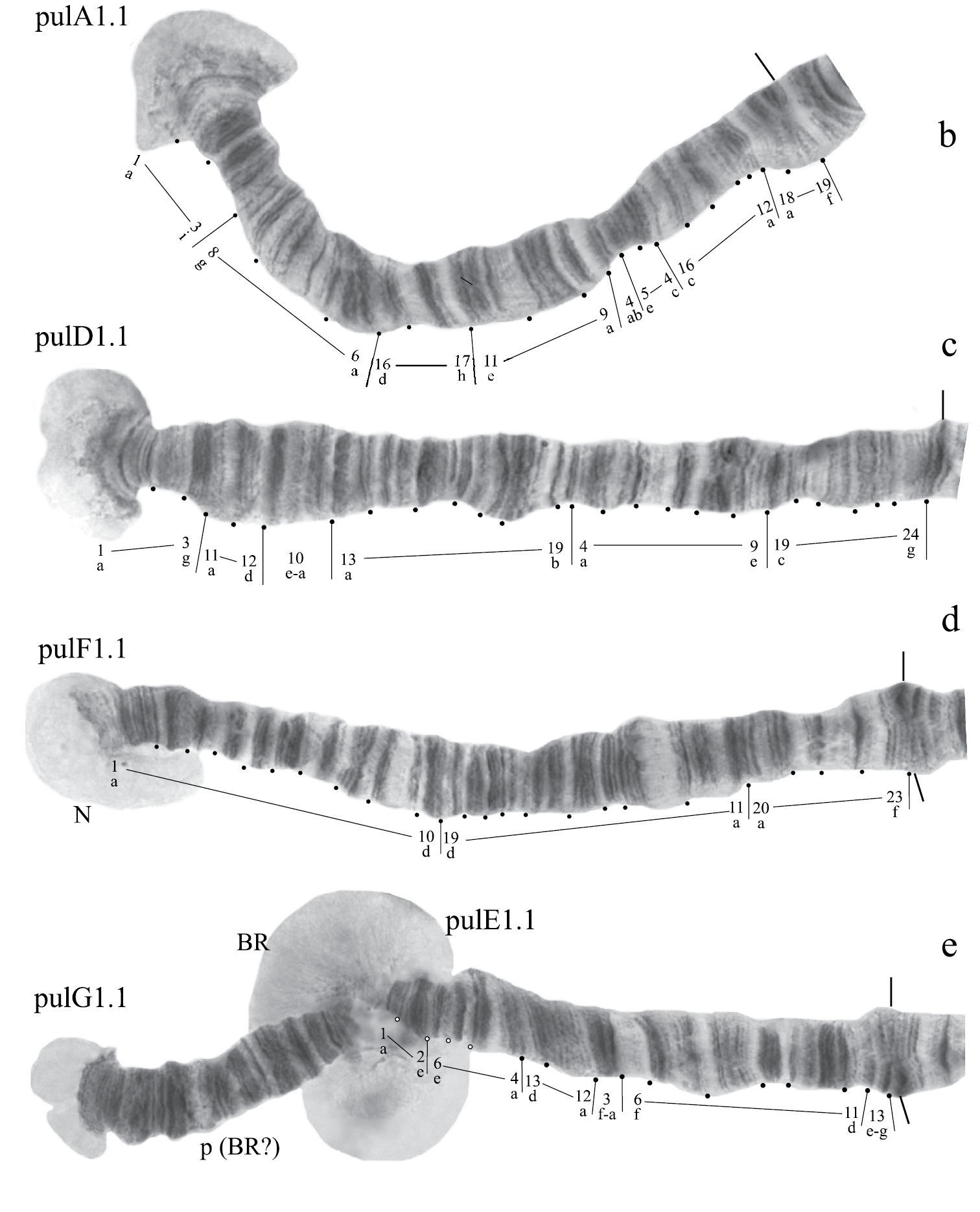
Karyotype of Chironomus prope pulcher. The designations are the same as in Fig. 1.
Homozygous banding sequences of Chironomus prope pulcher in arms A, D, E, F, and G.
Banding sequences (Fig. 5a, b-e).
Arm A (Fig. 5b) has the banding sequence pulA1, formed by four inversions from pigST
Arm B (Fig. 5a) not mapped, monomorphic. It has a sequence pulB1. The common BR is well developed.
Arm C (Fig. 5a) not mapped. It has two banding sequences pulC1 and pulC2 differing by a simple inversion, which involved practically the whole central part of arm C.
Arm D (Fig. 5c) has the sequence pulD1, formed by five inversion steps from pigST
Arm E (Fig. 5e) has the banding sequence pulE1, formed by three inversion steps from pigST.
Arm F (Fig. 5d) has the sequence pulF1, formed by one simple inversion from pigST.
The characteristic of arm F in Chironomus prope pulcher is the presence of the nucleolus at the telomeric end, which is a rare event among Chironomus species.
Arm G (Fig. 5e) is joined with arm E. There is large Balbiani ring near the site of fusion, and a small Balbiani ring or puff in the center of arm G. A small nucleolus is possibly developed at the telomeric end of arm G.
In total, eight banding sequences were recorded in the Chironomus prope pulcher banding sequence pool. All of them are endemic for Ethiopia. There are no basic sequences.
long tubuli laterales on abdominal segment VII. Other characters -
two pools within a short distance, River Athi south of Nairobi, Kenya.
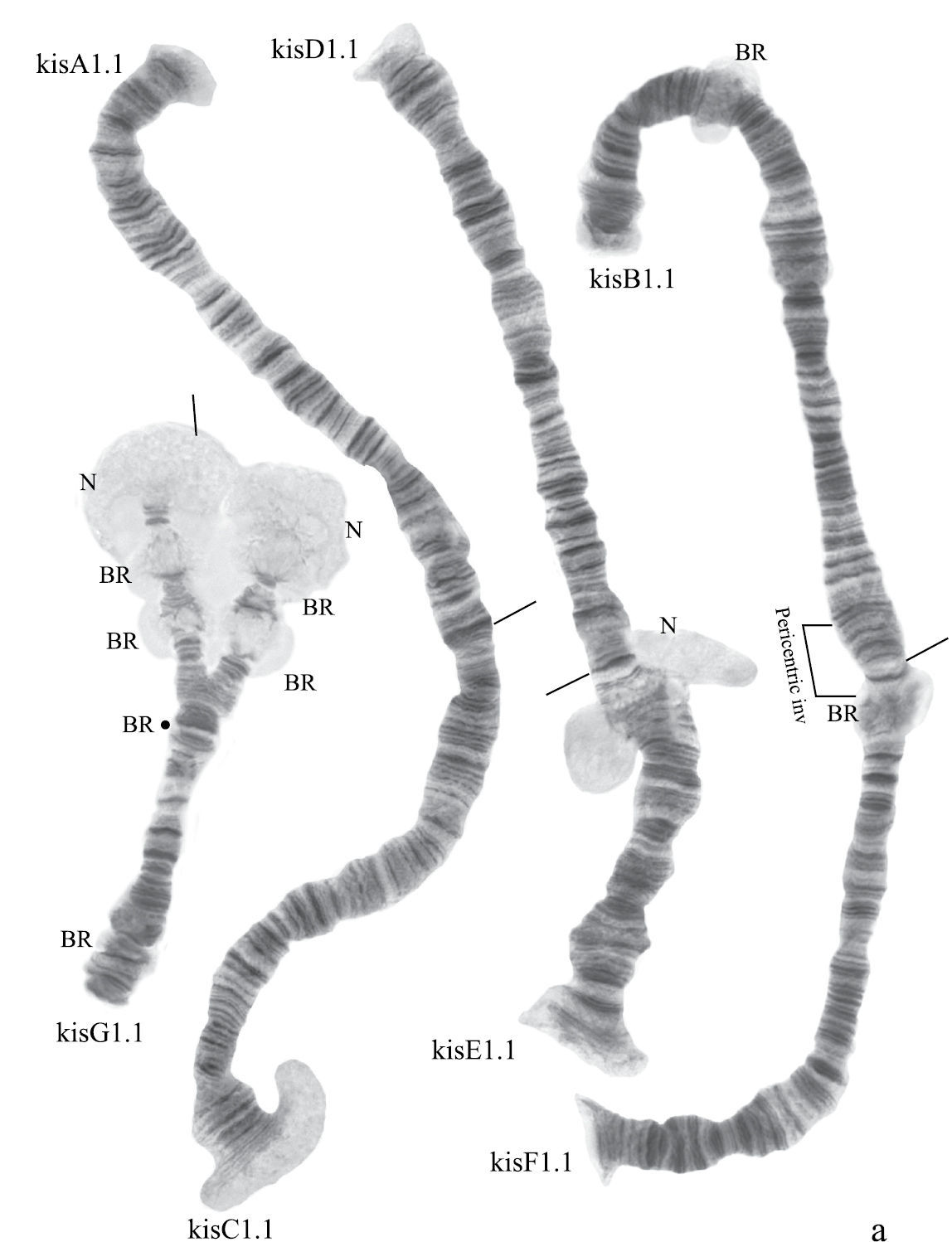
(Fig. 6a). Haploid number n=4, arm combination AC BF DE G (“parathummi”cytocomplex), centromeric bands not heterochromatinized, nucleoli in arms E and G, Balbiani rings in arms B and G. Chromosomal polymorphism was not recorded.
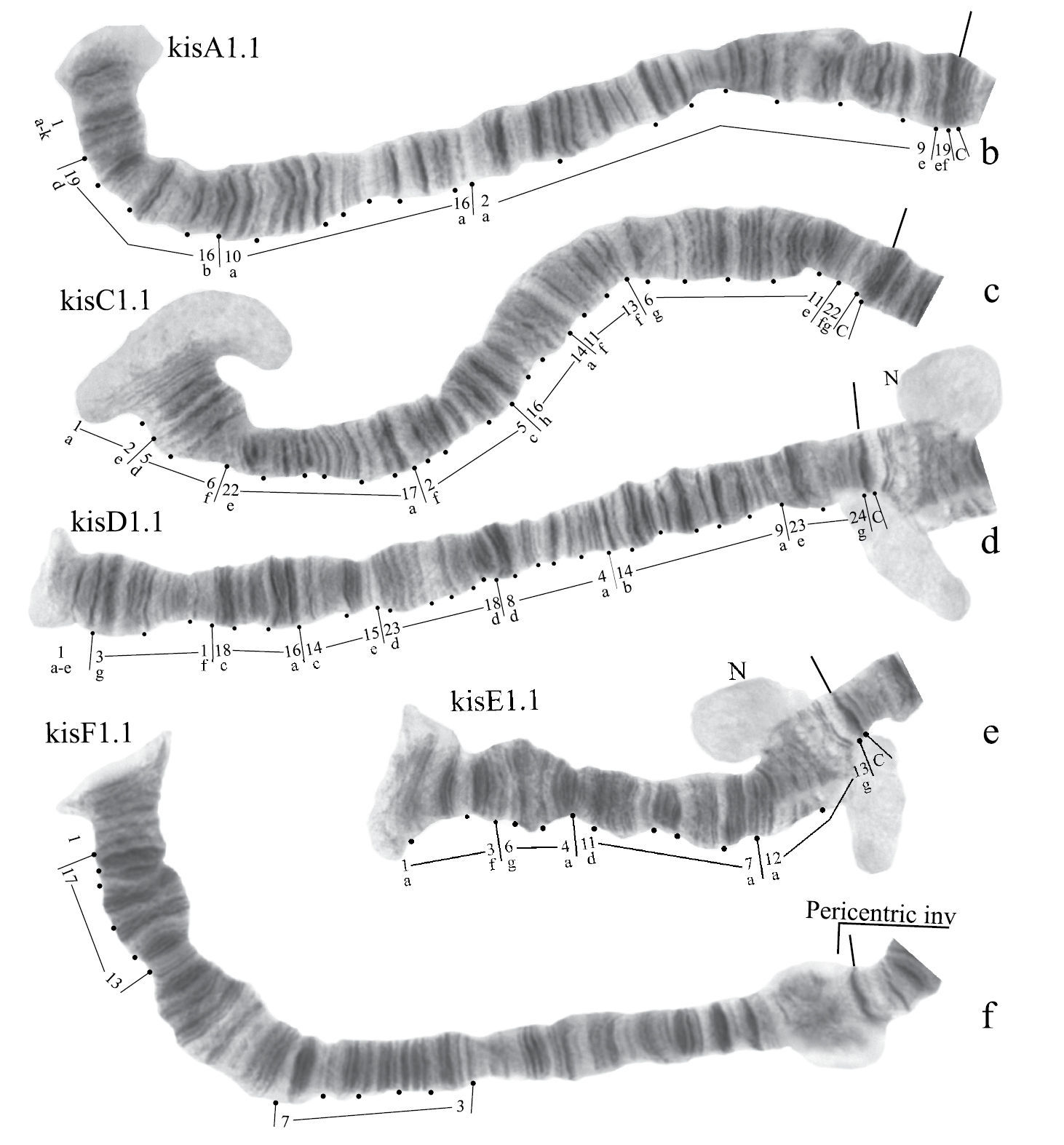
Karyotype of Chironomus sp. Kisumu.
Homozygous banding sequences of Chironomus sp. Kisumu in arms A, C. D, E and F.
Banding sequences (Fig. 6b-f)
Arm A (Fig. 6b) has the sequence kisA1, formed by 3 inversion steps from pigST.
Arm C (Fig. 6c) has the sequence kisC1, formed by 8 inversion steps from pigST.
Arm D (Fig. 6d) has the sequence kisD1, formed by 6 inversion steps from Chironomus piger ST:
Arm E (Fig. 6e) has the sequence kisE1, formed by two inversion steps from Chironomus piger ST
Presence of a nucleolus in region 13 in arm E is a great characteristic of the Chironomus sp. Kisumu karyotype.
Arm B (Fig. 6a) not mapped. It has one sequence – kisB1. The common BR is well developed.
Arm F (Fig. 6f) has the sequence kisF1. It was mapped only fragmentarily because of complex inversions in comparison with Chironomus piger ST. The presence of a large Balbiani ring situated just near the centromeric band is a characteristic of arm F in the Chironomus sp. Kisumu karyotype. There is pericentric inversion in the chromosome BF (Fig. 6a, f).
Arm G (Fig. 6a) is longer than usual in Chironomus species. There is a nucleolus and four Balbiani Rings on arm G. One of Balbiani Rings, noted by the black dot in Fig. 6a, was developed only in some cells of the salivary gland cells.
In total, seven Ethiopian endemic banding sequences are found in the sequence pool of Chironomus sp. Kisumu. All these sequences differ from Chironomus parathummi Keyl, 1961 sequences.
long tubuli laterales on abdominal segment VII.
near Victoria lake, Kenya.
Karyotypes of six African Chironomus species were studied. Four of these karyotypes were described for the first time (Chironomus sp. Nakuru, Chironomus formosipennis, Chironomus prope pulcher, Chironomus sp. Kisumu). Detailed photomaps of arms A, C, D, E, and F were presented, also for the first time, for Chironomus alluaudi, Chironomus transvaalensis, and Chironomus sp. Nakuru.
Among the species studied, three species (Chironomus transvaalensis, Chironomus prope pulcher, Chironomus
sp. Kisumu) have only endemic Ethiopian banding sequences in their
karyotypes, while cosmopolitan basic banding sequences were discovered
in the karyotypes of the other species, along with endemic sequences (Chironomus alluaudi, Chironomus sp. Nakuru, Chironomus formosipennis). The presence of these basic sequences indicates a relationship of African Chironomus species to Chironomus species from other continents before their separation (
The results on African species are relevant the problem
whether or not the chromosome arm combination of the “thummi”
cytocomplex is rare in Southern continents. At the moment, one species
in South America (Chironomus sp. Las Brisas, Wülker, Morath, 1989), one species in India (Chironomus javanus Kieffer, 1924), and two species in Australia (Chironomus javanus, Chironomus queenslandicus Martin, 2005) are known to have this “thummi” cytocomplex chromosome arm combination (
Earlier it was demonstrated (
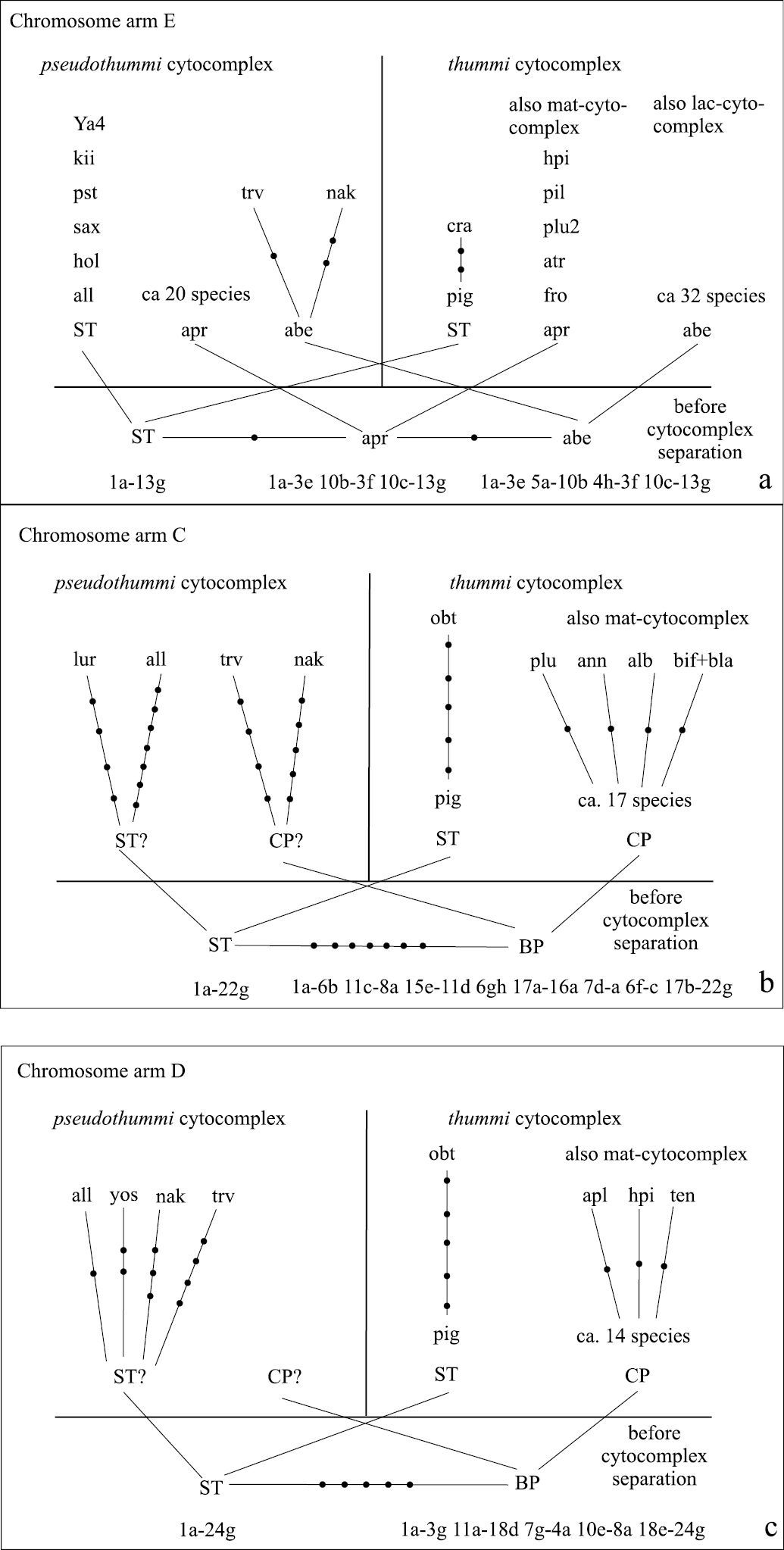
Relations of recent species and hypothetical “basic” species before separation of the cytocomplexes in arm E (a), arm C (b), and arm D (c). The data of
In arms C and D, an accumulation of species with the
identical sequences was previously observed only in the “thummi”
cytocomplex (
A great peculiarity of some African Chironomus karyotypes is the presence of large numbers of functionally active chromosome sites, especially Balbiani rings. For example 5 BRs were found in Chironomus transvaalensis (Figs 2a, 2g-i), 6 BRs in Chironomus sp. Nakuru (Fig. 3a). Most Chironomus species have two or three visible BRs since e.g. many species have the gene for BR4 but do not express it, and the number seen may also vary with developmental stage.
Travel of W.W. was sponsored by Deutsche Forschungsgemeinschaft. In Nairobi, family B. and R. Seelmann-Eggebert offered great hospitality and cooperation, Prof. Schneider and Prof. Kaib gave important informations. Prof. Dr. Vareschi, Drs. Reyer, Pater N. Cox, Oltmanns, d’Oliveira and J. Grunewald helped in collection of the larvae. Wülker’s family contributed encouragement, car driving and food. Many thanks to all of them.
The work was supported by the Presidium of the Russian Academy of Sciences under the program “The Origin and Evolution of Biosphere”, subprogram 2 project “The Role of Chromosome Rearrangements in Speciation and Evolution”; the program “Biodiversity and Gene Pool Dynamics”, subprogram “The Dynamics of Plant, Animal, and Human Gene Pools”, project “The Mechanisms of Genetic Variation of Diptera Populations and Species” and by the grant of RFBR No 09-04-01440.