(C) 2011 Ernani de Oliveira Mendes-Neto. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
The paper represents a comparative cytogenetic analysis of three populations of Hypostomus regani in Brazil.Two populations belong to the Upper Paraná River Basin and the third one, the karyotype of which is described for the first time, was probably introduced into the São Francisco River Basin through transposition from the Piumhi River. Karyotype features of populations of Hypostomus regani from the Piracicaba and Tietê River Basins are also discussed. The occurrence of Hypostomus regani in the São Francisco River Basin is reported for the first time here. The study also revealed distinct differences in the location of the Ag-NORs between the analyzed populations that enable individuals from the Piumhi River, Mogi-Guaçu River and Tietê River to be distinguished from one another. Thus, the data obtained indicate the possibility of geographic variation fixing different karyotypes even in the same basin of origin.
cytotaxonomy, karyotype diversification, rDNA
The family Loricariidae is the second most numerous among fish, with 716 species distributed among 96 genera (
Hypostomus Lacépède, 1803 is the largest genus of the armored catfish family Loricariidae, with approximately 120 nominal species (
In the present study, a comparative cytogenetic analysis was carried out on three different populations of Hypostomus regani (Ihering, 1905). Two populations are from the Upper Paraná River Basin and the other, the karyotype of which is described for the first time, was probably introduced into the São Francisco River Basin through the transposition of the Piumhi River. The aim was to investigate the karyotype in these populations, seeking chromosomal characters potentially important for understanding the taxonomy and biogeography of the species.
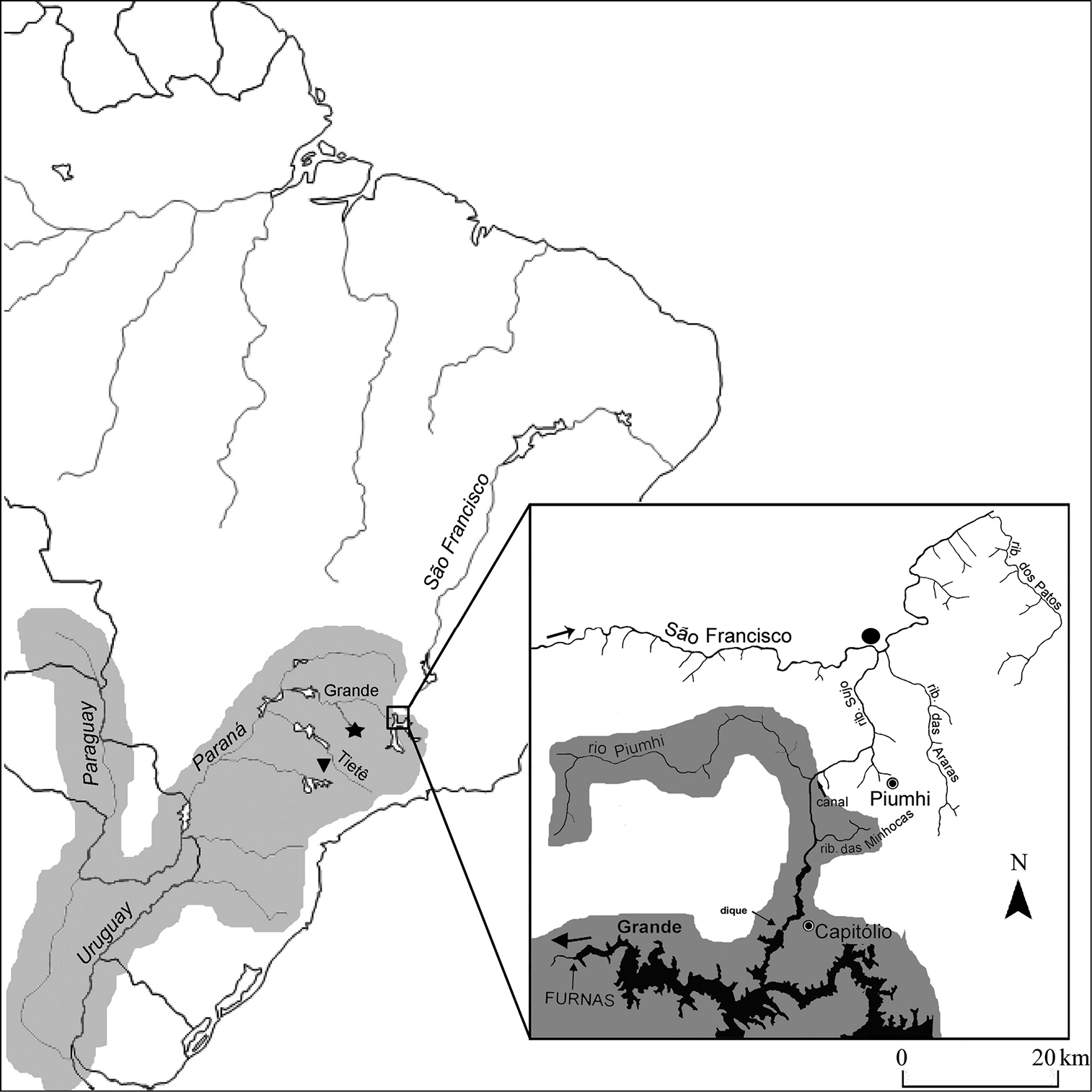
Material and methodsSixteen specimens of Hypostomus regani were examined (8 males and 8 females), collected from the mouth of the Piumhi River at the São Francisco River in the region of the municipality of Piumhi – MG, Brazil (20°20'31.0"S, 45°59'03.4"W, Alt.: 640 m), (Fig. 1, detail).According to C.H. Zawadzki (personal communication), Hypostomus regani is characterized as a species with a body covered by small, round, light-colored and generally well-defined spots. It has a high body and relatively long (narrow) head, large eyes and long dorsal fins, generally with rays reaching the spine of the adipose fin when adpressed. It has plates on the abdomen, except for very young specimens. Although distributed throughout the Paraná-Paraguay Basin, its type locality is the Piracicaba River in the state of São Paulo, Brazil.
Map of Brazil highlighting the large hydrographic basin of the Paraná-Paraguai system, area of natural distribution of Hypostomus regain.
Detail: area of the divider of the waters of the Upper Paraná and São
Francisco River Basins, altered by the transposition of the Piumhi
River (with original drainage to the Grande River in the Upper Paraná
Basin) to the Upper São Francisco Basin through an artificial channel
that links it to the Sujo River (tributary of the São Francisco River).
Star () and triangle (▼) indicate sampling sites for the Hypostomus regani populations studied by (
The specimens were identified and deposited in the Museu Nacional do Rio de Janeiro (MNRJ 32778; MNRJ 32782; MNRJ 32787). The collection authorization (number 472897) was granted by IBAMA [Brazilian Environmental Protection Agency] and the fishing license (number 091/07) was granted by the Instituto Estadual de Floresta de Minas Gerais, Brazil.
Karyotype data on the Hypostomus regani populations studied by
Chromosomal data of the Hypostomus regani populations; from
| Locality | 2n/FN | Formula | Ag-NOR | Ref. |
|---|---|---|---|---|
| Rio Mogi-Guaçu, Rio Mogi-Guaçu basin | 72/116 | 10m+20sm+42st/a | Multiple1 pair “a” large1 par “st” small | 1 |
| Rio Araquá, Rio Tiete basin | 72/116 | 12m+18sm+26st+16a | Multiple2 pairs “a” largies | 2 |
| Confluence of the Rio Piumhi with to Rio São Francisco, upper Rio São Francisco basin | 72/116 | 8m+16sm+20st+28a | Simple1 pair “st” large | 3 |
Chromosome preparations were obtained from cells from the anterior portion of the kidney, using in vivo treatment with colchicine (
Fluorescent in situ hybridization (FISH) was employed to locate ribosomal genes in the chromosomes. An 18S rDNA probe from the fish Prochilodus argenteus (Agassiz, 1829)(
Approximately 30 metaphases from each specimen were
analyzed in order to determine the modal diploid number (2n),
fundamental number (FN) and karyotype formula. The chromosomes were
identified based on the approach described by
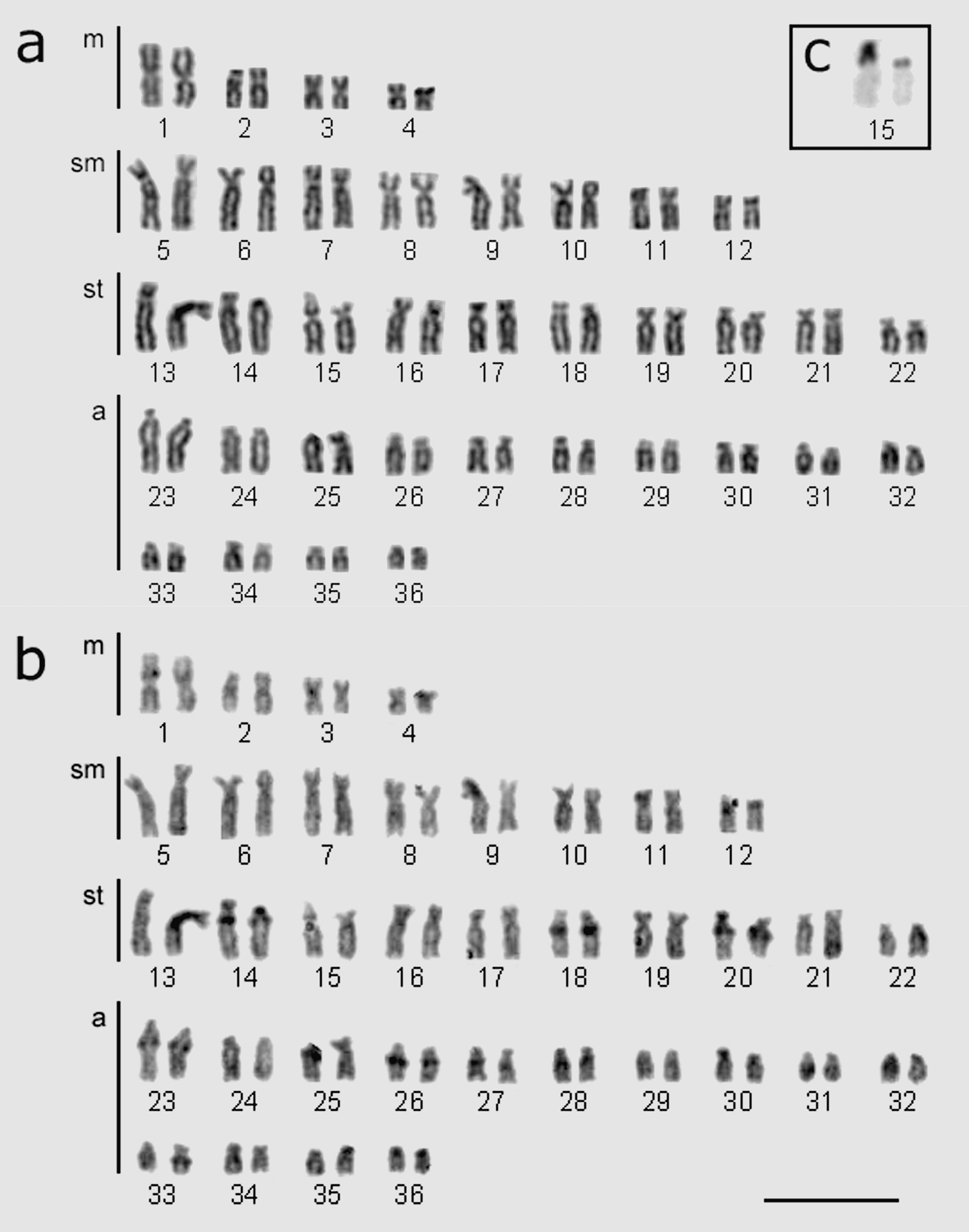
All the Hypostomus regani specimens analyzed in the present study had 2n = 72 chromosomes with a karyotype formula 8m+16sm+20st+28a. The number of arms was FN = 116 (Table 1, Fig. 2a).
Hypostomus regani karyotypes from the confluence of the Piumhi and São Francisco Rivers a chromosome stained with Giemsa and b sequentially labeled by C-banding; c box indicates chromosome pair labeled with silver nitrate locating the nucleolus organizer regions (pair no. 15). Bar = 10 µm.
Constitutive heterochromatin was distributed in small blocks (Fig. 2, b). The interstitial region of the long arms of subtelocentric chromosomes pairs 14, 18 and 20 as well as acrocentric pairs 23, 26, 27, 28 and 33 had quite evident blocks. Metacentric chromosome pair 1 had fainter labeling in the interstitial region of the short arm. Chromosome pairs 13, 15 and 36 had heterochromatic blocks in the centromeric region. Heterochromatin was located in the telomeric region of the long arm in chromosome pair 31. Nucleolar organizing regions (NORs) labeled by silver nitrate were only evident in the short arm of subtelocentric chromosome pair 15 (Fig. 2, c and 4, a). All cells analyzed exhibited heteromorphism in relation to the size of the Ag-NORs.
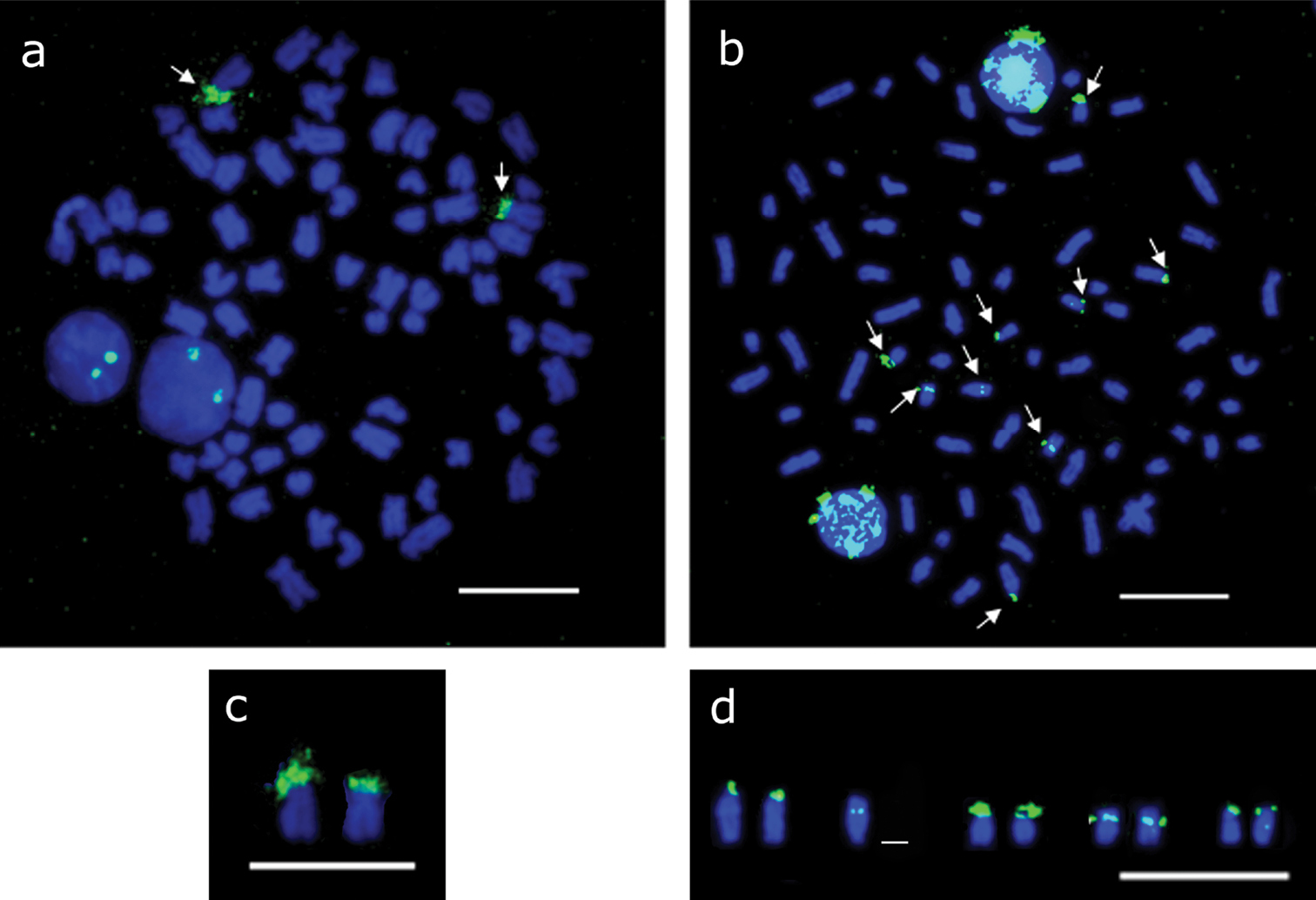
Fluorescent in situ hybridization confirmed the presence of 18S rDNA coinciding with the Ag-NORs as well as the size heteromorphism of the sites (Figs 3, a-c). The 5s rDNA sites were located in four chromosome pairs: in the terminal region of the short arm of two acrocentric pairs; in the centromeric region of one submetacentric pair; and on another chromosome with no evident homologous labeling (Figs 3, b-d).
Mitotic metaphases in Hypostomus regani from the confluence of the Piumhi and São Francisco Rivers submitted to fluorescent in situ hybridization a showing two 18S rDNA sites (arrows) b nine 5S rDNA sites (arrows) c chromosomes bearing 18S rDNA sites d chromosomes bearing 5S rDNA sites. Bar = 10 µm.
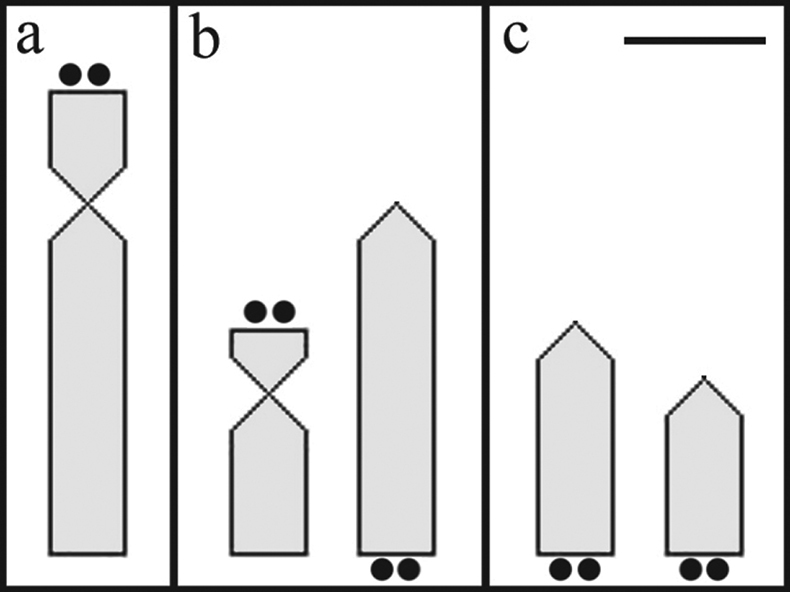
Idiogram of chromosomes bearing nucleolus organizer regions labeled by silver nitrate, comparing Hypostomus regani populations from the Piumhi River in the Upper São Francisco Basin a and Mogi Guaçu b and Tietê c Rivers in the Upper Paraná Basin. Bar = 1 µm.
The Piracicaba River, which is a tributary of the Upper Paraná River, is the type locality of Hypostomus regani, although the natural distribution of the species is related to the Paraná, Paraguay and Uruguay River Basins (
Karyotype differences in natural fish populations that
inhabit the same hydrographic basin have been found, for instance, in
the genus Astyanax Baird et Girard, 1854. E.g. there are at least three different cytotypes of Astyanax prope fasciatus (Cuvier, 1819) living in sympatry in the Upper Tibagi River, which is a tributary of the Paraná River (
In a previous study,
Especially regarding the species Hypostomus regani,
we can highlight the location of nucleolus organizer regions (NORs) as
a variable inter-population character. The results reveal distinct
patterns of chromosome types and location that enable the Hypostomus regani
population in the Piumhi River of the São Francisco Basin to be
distinguished from the populations in the Mogi-Guaçu and Tietê Rivers
analyzed by (
The results obtained with the chromosomal location of 5S gene do not currently allow any evolutionary inferences. However this cytotaxonomical marker may be important in future studies, especially regarding the number and location of this class of ribosomal DNA in Hypostomus.
Besides the identification of a population of Hypostomus regani belonging to the native ichthyofauna of the Piumhi River basin, originally founded from the ichthyofauna of the Upper Paraná River, we also verified the introduction of an exotic species in the São Francisco River Basin, with unpredictable consequences for the homeostasis of this environment.
The authors are grateful to the Instituto Estadual de Floresta de Minas Gerais (License number 091/07) and IBAMA (Instituto Brasileiro do Meio Ambiente IBAMA/MMA/SISBIO license number: 472897). This work was supported by FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo, Processo: 06/54290-6), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior).