(C) 2011 D.B. Carvalho. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Chromosomes of Ommexecha virens and Descampsacris serrulatum (Ommexechidae) were analyzed through conventional staining, C-banding, base specific fluorochromes, silver nitrate impregnation (AgNO3), and fluorescent in situ hybridization (FISH) with probe for 45S rDNA. The two species presented diploid number 2n= 23, X0 in males and acrocentric autosomes, except the pair one that presented submetacentric morphology. The X chromosome has distinct morphology in the two analyzed species, being a medium acrocentric in Ommexecha virens and large submetacentric in Descampsacris serrulatum. The C-banding revealed pericentromeric blocks of constitutive heterochromatin (CH) in all the chromosomes of Descampsacris serrulatum. For Ommexecha virens it was evidenced that the blocks of CH are preferentially located in the pericentromeric area (however some bivalents presents additional blocks) or in different positions. The staining with CMA3/DA/DAPI showed GC rich CH blocks (CMA3+) in some chromosomes of the two species. The nucleolar organizer regions (NORs) were located in the bivalents L2, S9, S10 of Ommexecha virens and M5, M6, M7, S11 of Descampsacris serrulatum. The FISH for rDNA showed coincident results with the pattern of active NORs revealed by AgNO3. This work presents the first chromosomal data, obtained through differential cytogenetics techniques in Ommexechidae, contributing to a better characterization of karyotypic evolution for this grasshopper family.
FISH, grasshopper, Ommexechidae, rDNA, Ommexecha virens, Descampsacris serrulatum
The family Ommexechidae is endemic to South America, constituted by two subfamilies (Aucacrinae and Ommexechinae) and includes about 12 genera and 30 species. Descampsacris is a monotypic genus represented by Descampsacris serrulatum. On the other hand, the genus Ommexecha comprises seven species, Ommexecha apolinari, Ommexecha brunneri, Ommexecha germari, Ommexecha giglio-tosi, Ommexecha gracilis, Ommexecha macropterum and Ommexecha virens. This genus has a wide geographical distribution being found from the Andes to Caribbean. Among the described species, Ommexecha virens presents great morphologic and chromatic variability (
Differential cytogenetic staining in grasshoppers of the families Acrididae and Romaleidae, from Neotropical Region, has shown a variability in distribution pattern and qualification of the CH through the C-banding and base specific fluorochromes Chromomycin A3 (CMA3) and 4’-6’-diamidino-2-fellindol (DAPI) staining. Distinct nucleolus organizing regions (NORs) locations, through the silver nitrate impregnation (AgNO3) and fluorescent in situ hybridization (FISH), have been observed in these species (
In this work chromosomes of Ommexecha virens and Descampsacris serrulatum were analyzed through conventional staining, C-banding, base specific fluorochromes, impregnation with AgNO3 and FISH with probe of 45S rDNA. The karyotypic patterns obtained contributed to a better understanding about chromosomal evolution in the family Ommexechidae.
Material and methodsIn this work 36 males of Ommexecha virens (Thunberg, 1824) and 11 of Descampsacris serrulatum (Serville, 1831) were analyzed. The species studied were collected in different areas in the states of Pernambuco and Bahia in the Northeast Region of Brazil (Table 1). The testes were fixed in ethanol and acetic acid 3:1. The cytological preparations were obtained through squashing of testes follicles. For conventional analysis the slides were stained with lacto acetic orcein 2%. For the C-banding the technique of
The silver nitrate impregnation (AgNO3) was done according to
Fluorescent in situ hybridization (FISH) was performed according to
The cells submitted to the fluorochromes and FISH were captured through the image capture system Cytovision coupled to the microscope Olympus BX51. For the other methods the cells were photographed in microscope Leica, using a film Kodak Imagelink, Wing 25. The figures were mounted with the use of the program Corel Draw Graphics Suite 12 software.
Species, localities of collections, geographical coordinates and number of analyzed individuals.
| Species | Localities | Coordinates | No. of analyzed individuals |
|---|---|---|---|
| Ommexecha virens (Serville, 1831) | Buíque (PE) | 8°37'23"S, 37°9'12"W | 8 |
| Itamaracá (PE) | 7°44'52"S, 34°51'19"W | 9 | |
| Sobradinho (BA) | 9°27'19"S, 40°49'24"W | 6 | |
| Rio de Contas (BA) | 13°34'44"S, 41°48'41"W | 7 | |
| Itaberaba (BA) | 12°31'39"S, 40°18'25"W | 6 | |
| Descampsacris serrulatum (Thunberg, 1824) | Rio de Contas (BA) | 13°34'44"S, 41°48'41"W | 6 |
| Andaraí (BA) | 12°48'26"S, 41°19'53"W | 3 | |
| Mucugê (BA) | 13°0'19"S, 41°22'45"W | 2 | |
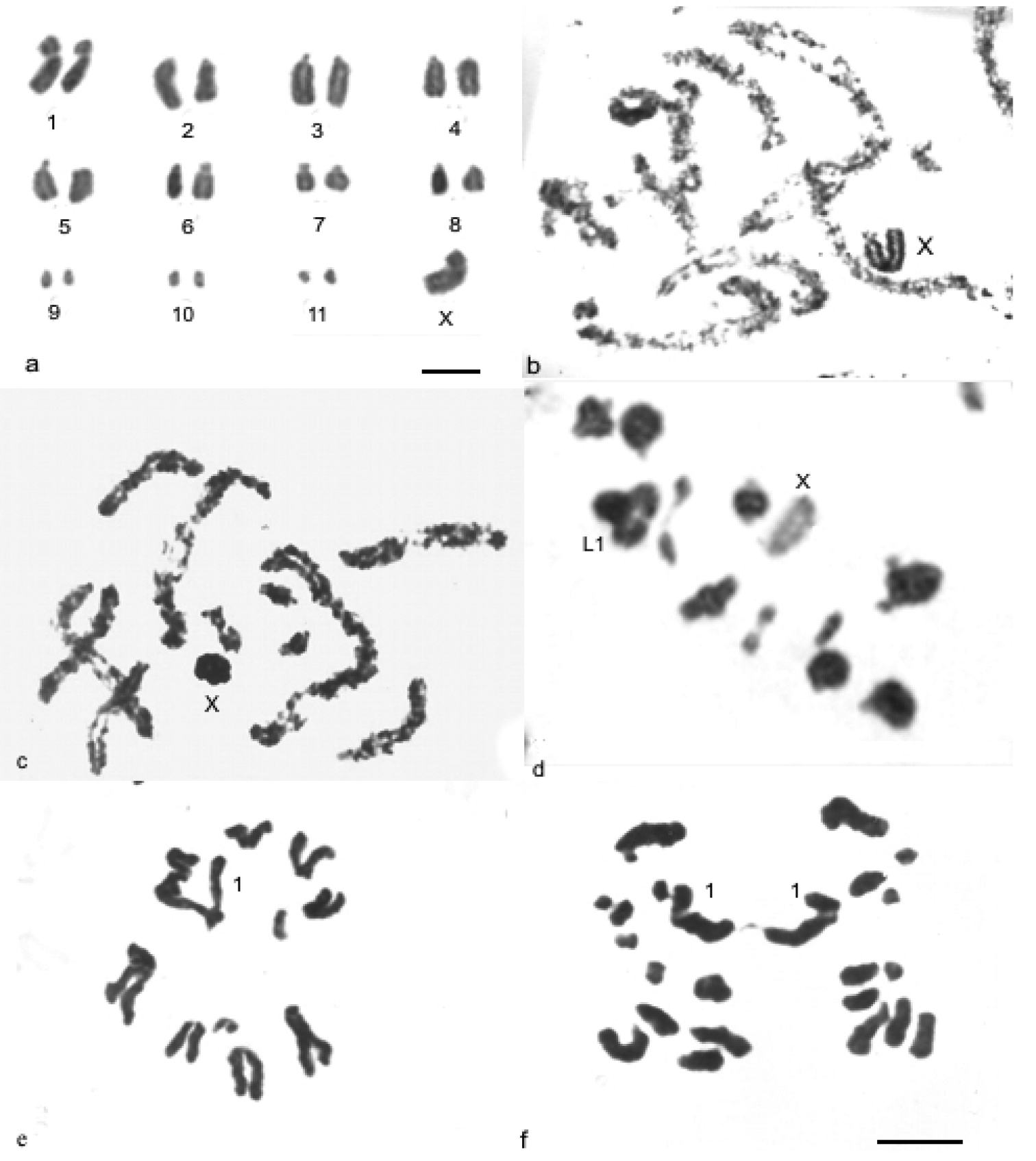
Descampsacris serrulatum and Ommexecha virens demonstrate similar karyotypes with diploid number 2n= 23, X0 in the males and acrocentric autosomes, except the pair one that presented submetacentric morphology (Fig. 1, a-f). The autosomes were arranged according to their size as three large (L1-L3), five medium (M4-M8) and three small (S9-S11). Descampsacris serrulatum has one large X chromosome with submetacentric morphology (Fig. 1, a, b), while in Ommexecha virens the X is of medium size and acrocentric (Fig. 1, c, d). During the prophase I of the two species the X chromosome was heteropycnotic positive until the diplotene, being that behavior reverted in the metaphase I (Fig. 1, b-d).
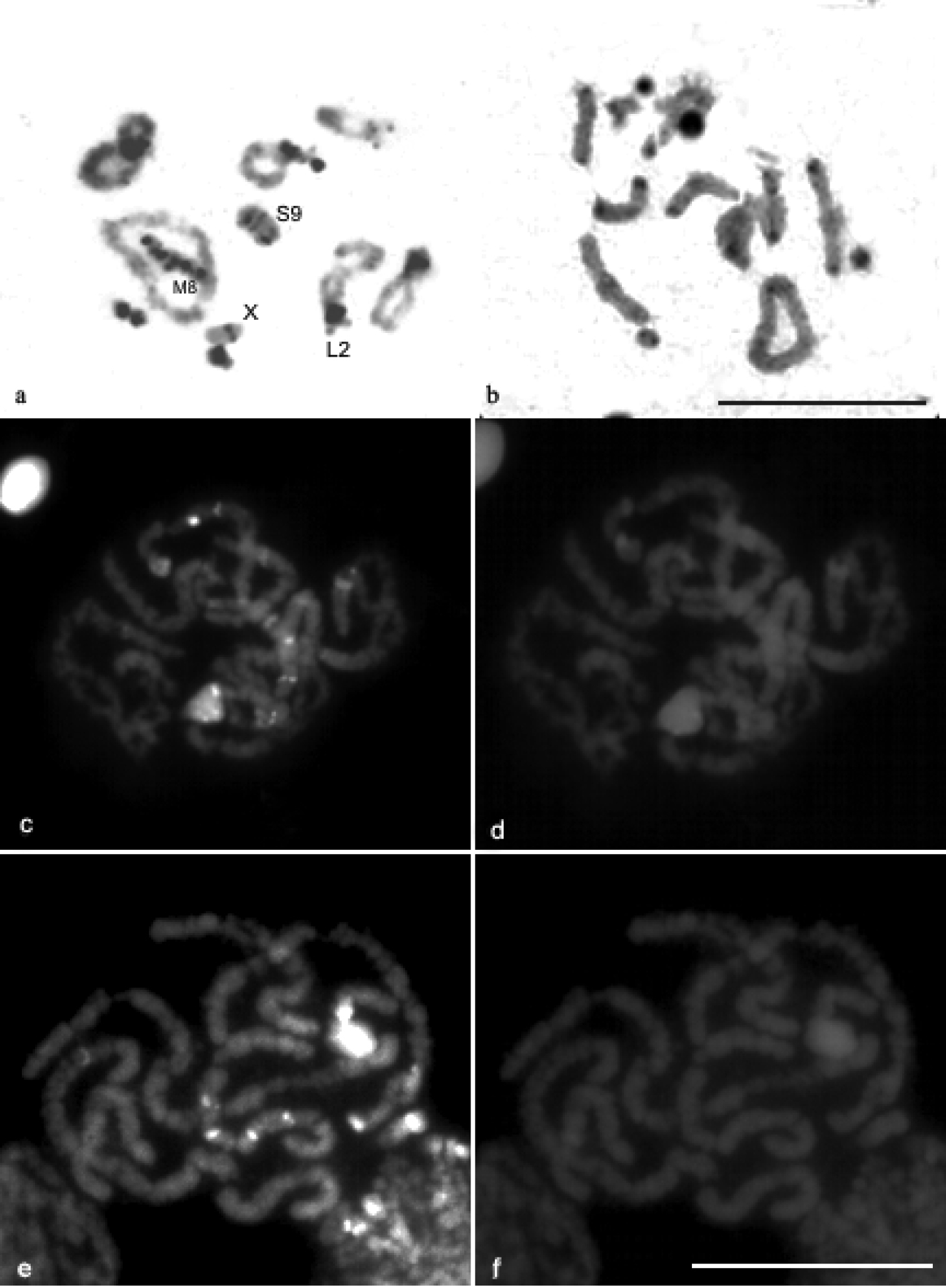
C-banding revealed distribution of constitutive heterochromatin (CH) in the pericentromeric region of all chromosomes of Descampsacris serrulatum (Fig. 2, b). In Ommexecha virens has blocks of CH preferentiality in the pericentromeric region, however some bivalents demonstrate additional blocks in different positions, such as L2 with proximal block, besides the pericentromeric; M8 showed intercalation of eu-heterochromatin along its extension; S9 presented a large block of CH extending from the pericentromeric to proximal region. Moreover the X chromosome showed proximal block (Fig. 2, a).
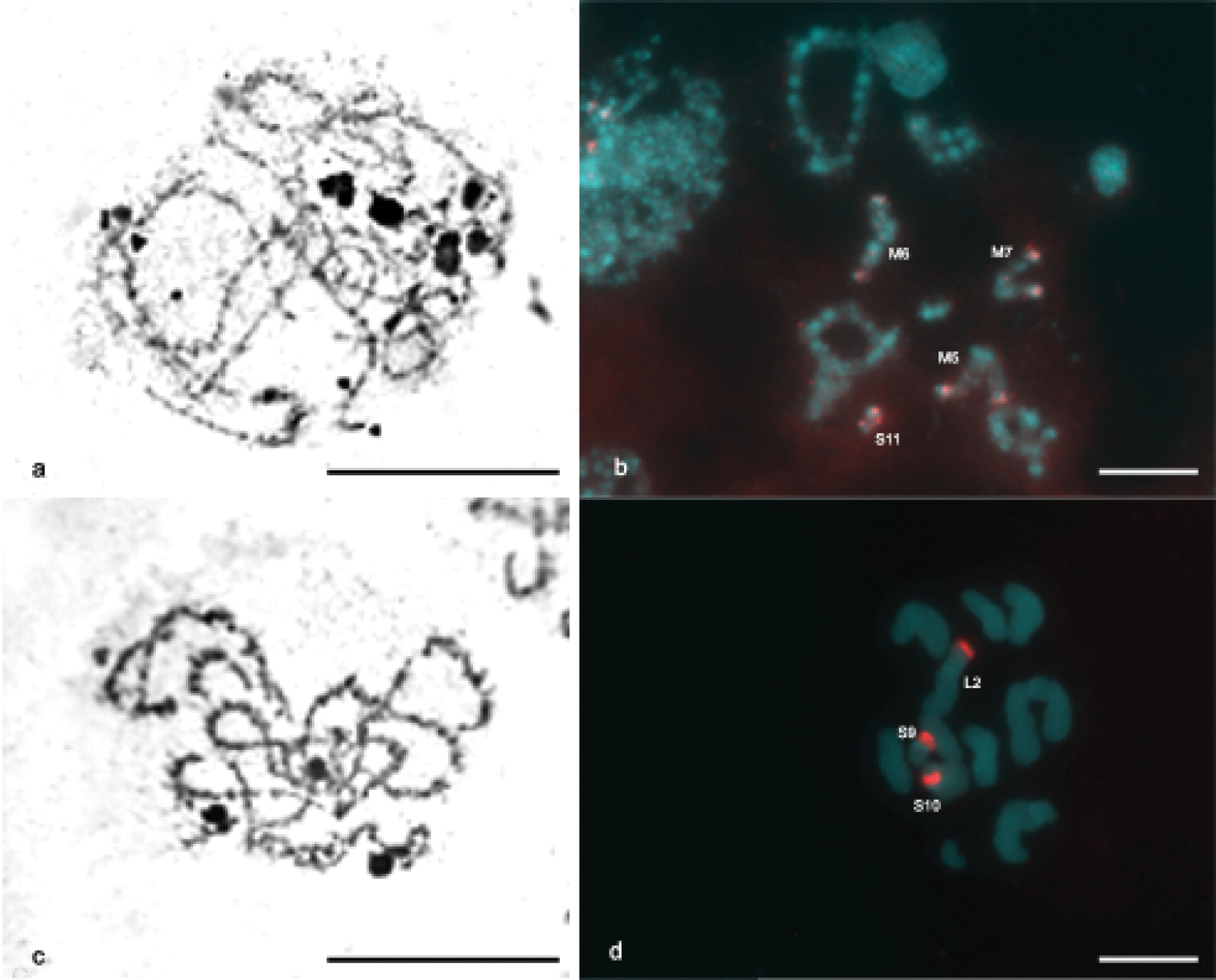
CMA3/DA/DAPI staining revealed blocks CMA3+ in some chromosomes of the karyotype of Ommexecha virens, as well as of Descampsacris serrulatum (Fig. 2, c, e). However, no AT positive blocks were detected in these two species (Fig. 2, d, f). Impregnation with AgNO3 identified nucleolus organizing regions (NORs) active in the two analyzed species. In Descampsacris serrulatum, NORs are located in four autosomal bivalents (M5, M6, M7 and S11) and in Ommexecha virens in three (L2, S9 and S10) (Fig. 3, a, c).
FISH with probe of 45S rDNA was used in meiotic cells of Descampsacris serrulatum and Ommexecha virens allowing the precise identification of rDNA sites observed by the impregnation AgNO3. In Descampsacris serrulatum it was observed that in four autosomal pairs the sites of rDNA are located in the pericentromeric regions (Fig. 3, b), besides the bivalent S11 presented a large sign of the hybridization, occupying about 2/3 of the chromosome length. In Ommexecha virens it was revealed that the bivalents L2 and S9 possess pericentromeric sites and the S10 proximal (Fig. 3, d).
Conventional staining in mitotic and meiotic cells of Descampsacris serrulatum a, b and Ommexecha virens c, f. a spermatogonial metaphase b and c pachytene d metaphase I e metaphase II f anaphase II. Bar= 5µm.
C-banding pattern in diplotene cells of Ommexecha virens a and Descampsacris serrulatum b Staining CMA3/DA/DAPI in pachytene cells of Descampsacris serrulatum c CMA3 d DAPI and Ommexecha virens e CMA3 f DAPI. Bar= 5 µm.
Impregnation with silver nitrate a, c and FISH with probe of 45S rDNA b, d. Zygotene and diakinesis of Descampsacris serrulatum a, b and zygotene and pachytene of Ommexecha virens c, d. In b, d are indicated the pairs containing the rDNA sites. Bar= 5 µm.
In spite of the wide distribution of the Ommexechidae in South America, the cytogenetic studies in this family are scarce and based mainly on conventional method (
The pericentromeric pattern of distribution of constitutive heterochromatin observed in Descampsacris serrulatum and Ommexecha virens is quite common for the superfamíly Acridoidea. This pattern has also been described for several species of the Neotropical Region, belonging the families Acrididae and Romaleidae (
The pattern of qualification of CH visualized by the staining CMA3/DA/DAPI in Descampsacris serrulatum and Ommexecha virens showed blocks of positive CMA3 in some chromosomes of the karyotype. Similar patterns with the presence of CMA3+ blocks in some chromosomes were also described in Belosacris coccineipes, Cornops aquaticum, Stenopola dorsalis, Stenacris xantochlorae and Tucayaca parvula (Acrididae) (
The references on the using of the AgNO3 impregnation and fluorescent in situ hybridization (FISH) for major rDNA show that in grasshoppers as a whole and in species belonging to the families Acrididae and Romaleidae, NORs are more frequently found in one or two autosomal bivalents (
The differential pattern of distribution of NORs observed in Descampsacris serrulatum and Ommexecha virens, could be explained by a probable amplification and dispersion of sites of rDNA (18S, 28S, 5.8S), leaving of an ancestral condition, in which a single autosomal pair would be NORs bearer. On the other hand, there is no coincident NORs bearing pairs among the two species and the possibility of different origins for the pattern of NORs in Ommexechidae can not be discarded.
We thank Dr. Carlos Salvador Carbonell of the University of Montevideo, Uruguay, for the taxonomic identification of the studied species and sending of bibliographical materials, to Prof. Alejo Mesa for the sending of bibliographical materials, Diogo Cabral-de-Mello for the critical reading of this paper and Cirlene Maria da Silva for the technical support. This research was supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico).