(C) 2011 Atilla Arslan. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Thekaryotype, C-banding, and nucleoar organizer regions (NORs) of eight specimens ofKonya wild sheepfrom Turkey were examined. The complement included six large metacentric autosomes, 46 acrocentric autosomes of decreasing size, a medium-sized acrocentric X chromosome, and a small bi-armed Y chromosome (the diploid chromosome number 2n=54, the number of autosomal arms NFa=58, the number of chromosome arms NF=61). G-banding allowed reliable identification of all the chromosome pairs and the pairing of homologous elements. All the autosomes possessed distinct centromeric or pericentromeric C-positive bands. The X chromosome had a pericentromeric C-positive band, and the Y chromosome was entirely C-heterochromatic. The NORs were located in the terminal regions of the long arms of three metacentric and two acrocentric autosomes. The karyotype of the Konya wild sheep and its banding patterns are quite similar to chromosome complement reported in domestic sheep and European mouflon.
chromosomes, Ovis, sheep domestication, Turkey
The systematics of the genus Ovis
(Linnaeus, 1758) is rather complicated and there are different
opinions in respect of the number of species and the actual species
borders (
The domestic sheep and its wild ancestors are sometimes considered conspecific, in which case the name Ovis orientalis has generally been used, although some authors use the name Ovis aries for both wild species and its domestic descendants (
Chromosome research has contributed significantly to
understanding of the evolutionary divergence among sheep taxa, and
different chromosome numbers were found in individual geographic
populations. The European mouflon and wild sheep from western
Palaearctic (Ovis orientalis)
have the same diploid number of 54 chromosomes as the domestic sheep.
In the Iranian populations of urial sheep the diploid number of 58
chromosomes was recorded, and hybrid zones with polymorphic karyotype
were found in the areas of contact with Ovis orientalis (
The karyotype of the Konya wild sheep was studied by
We examined eight specimens of Konya wild sheep including
four females and four males. All the studied animals originated from
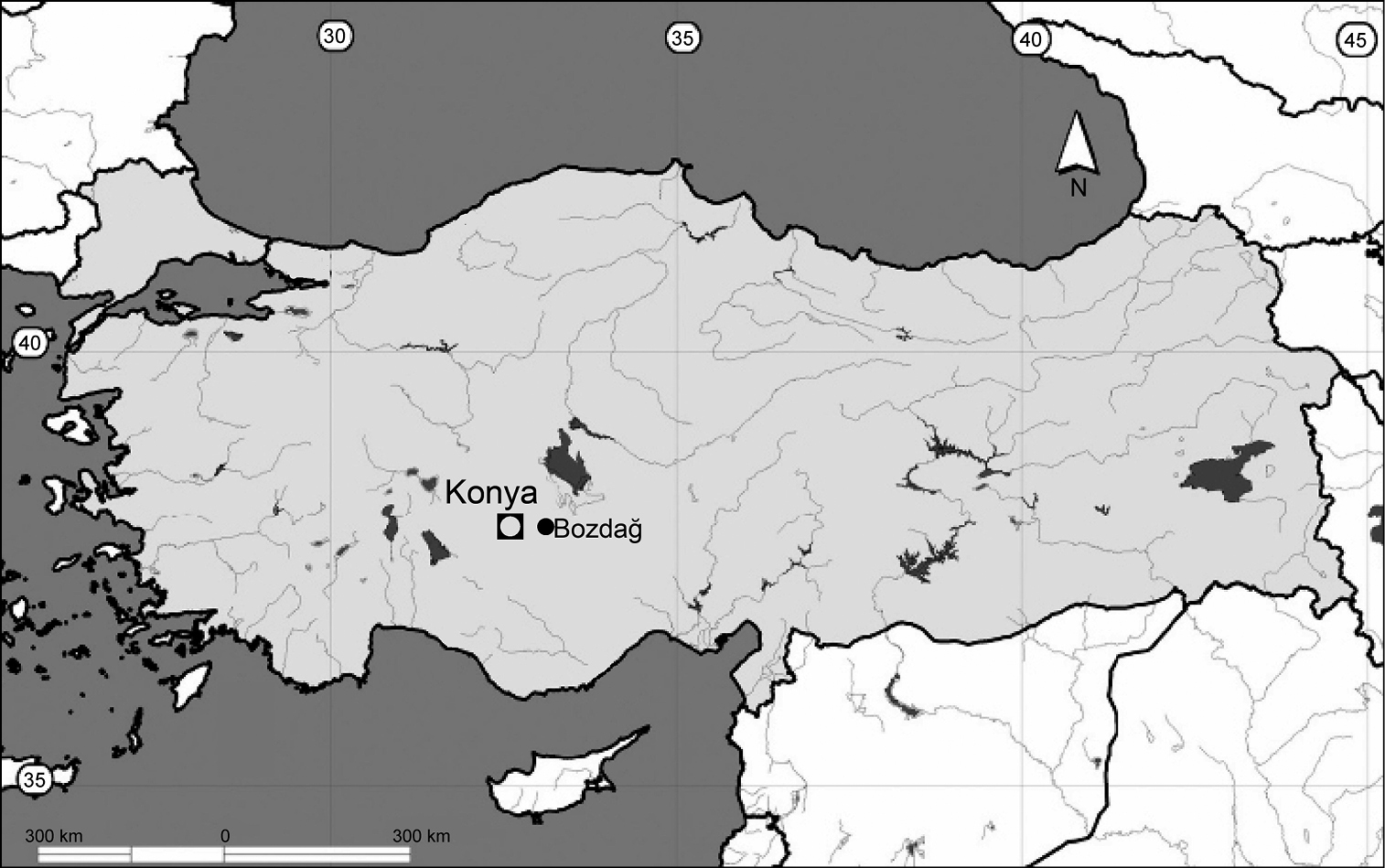
Bozdağ in the Konya province, Turkey (Fig. 1).
Chromosomal preparations were made using peripheral blood culture for
72 hr at 37oC in medium supplemented with 15% fetal calf serum and 0.024
% phytohaemagglutinin (
Collecting locality in the province of Konya, Anatolia, Turkey (●).
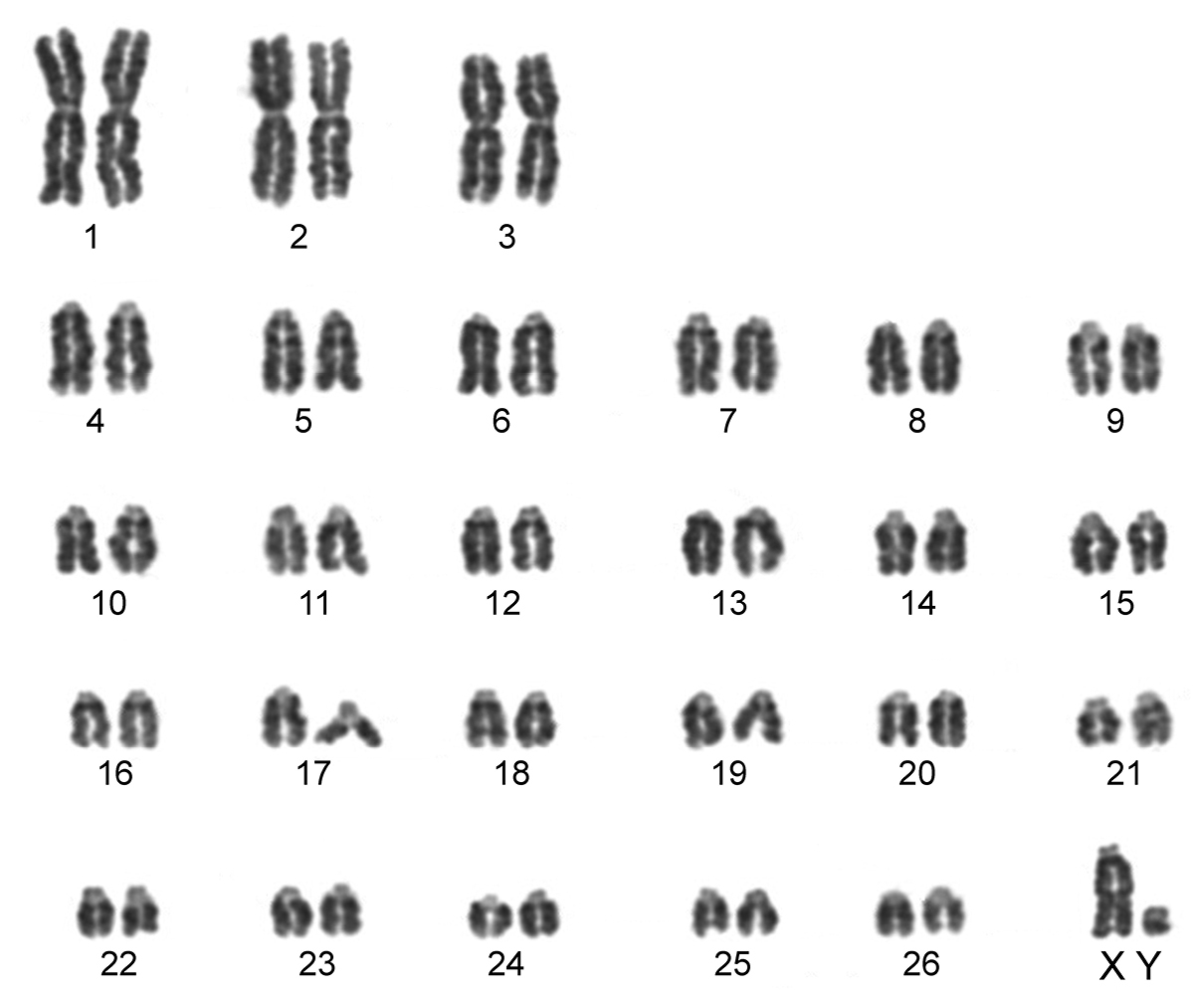
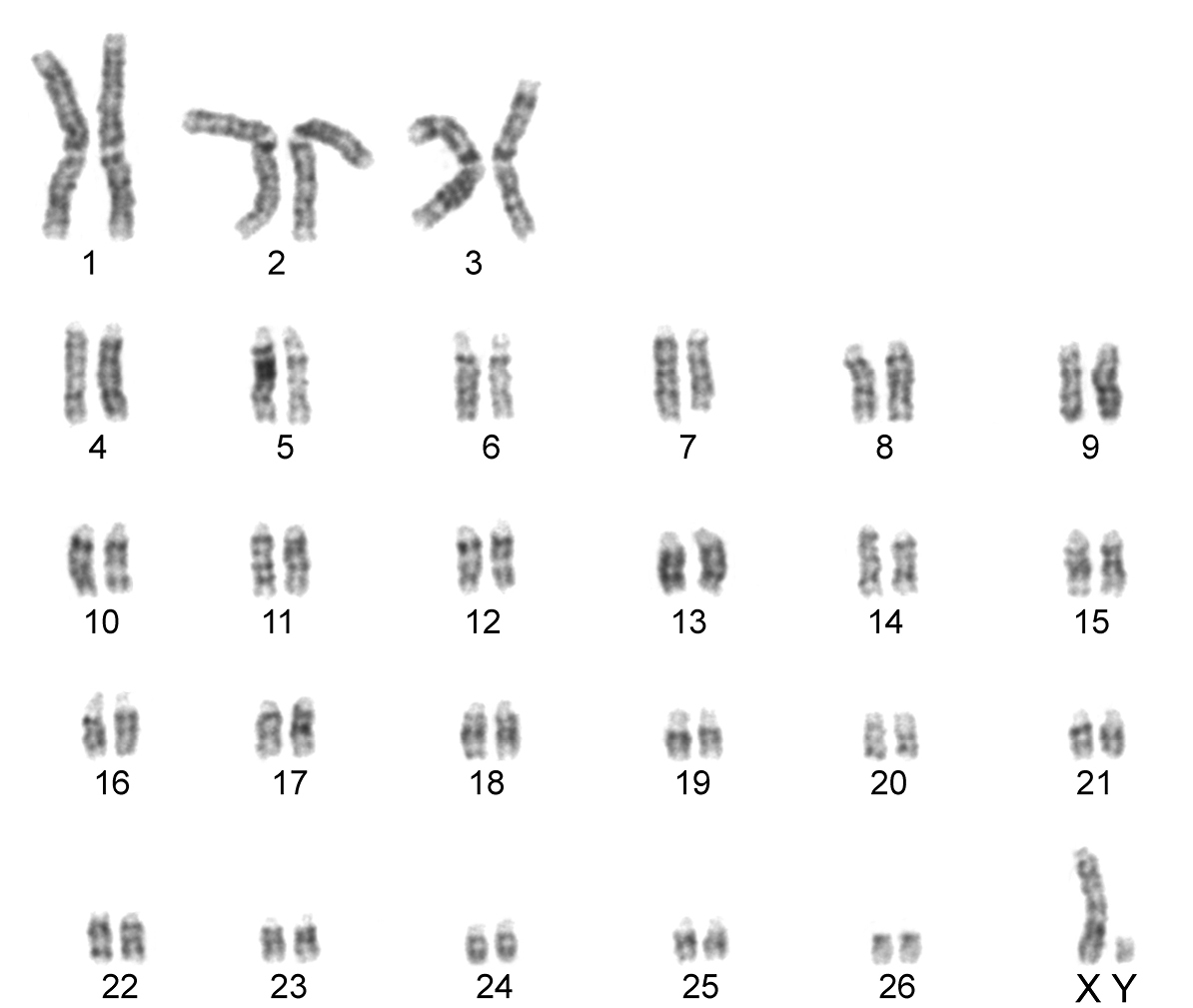
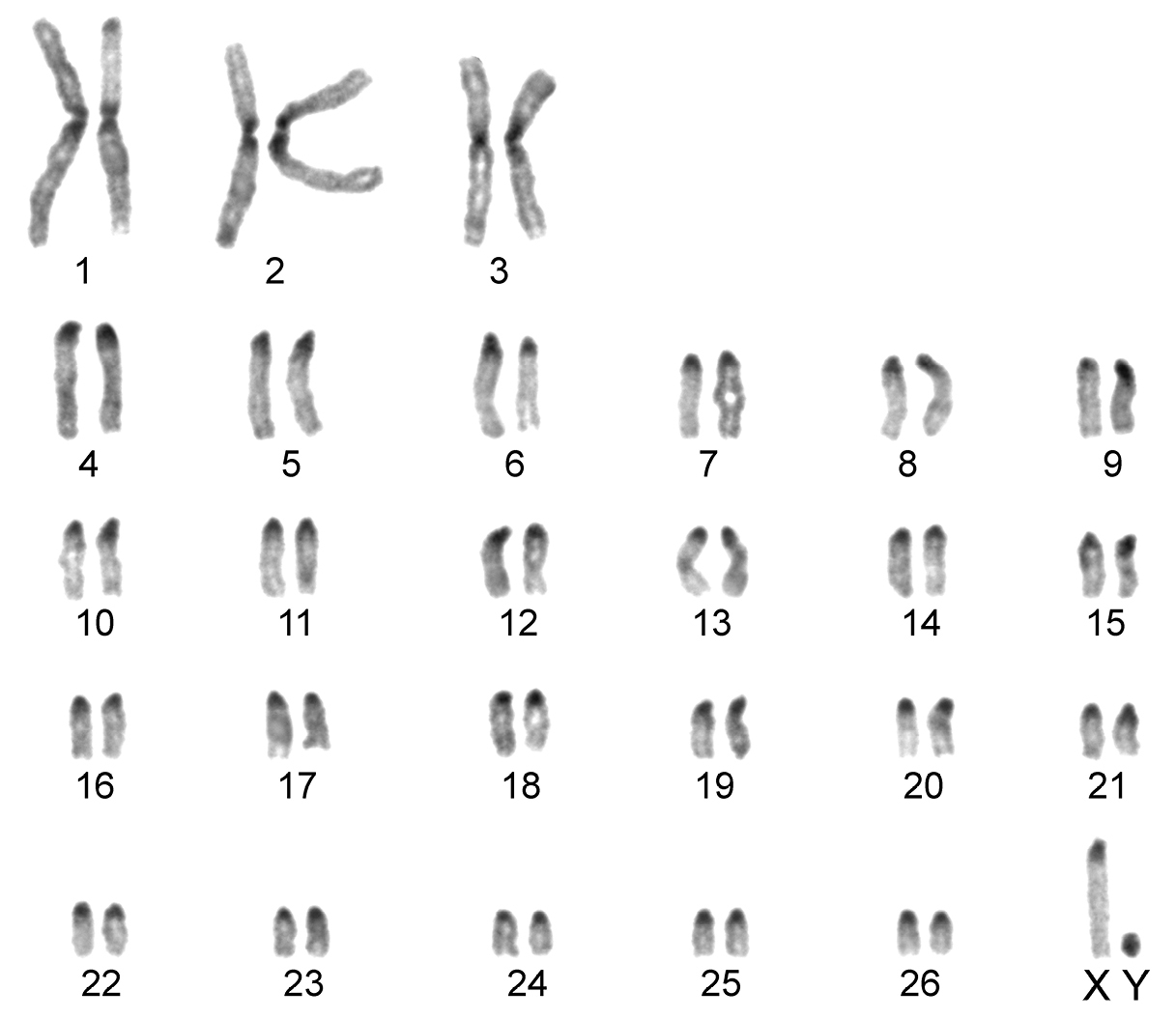
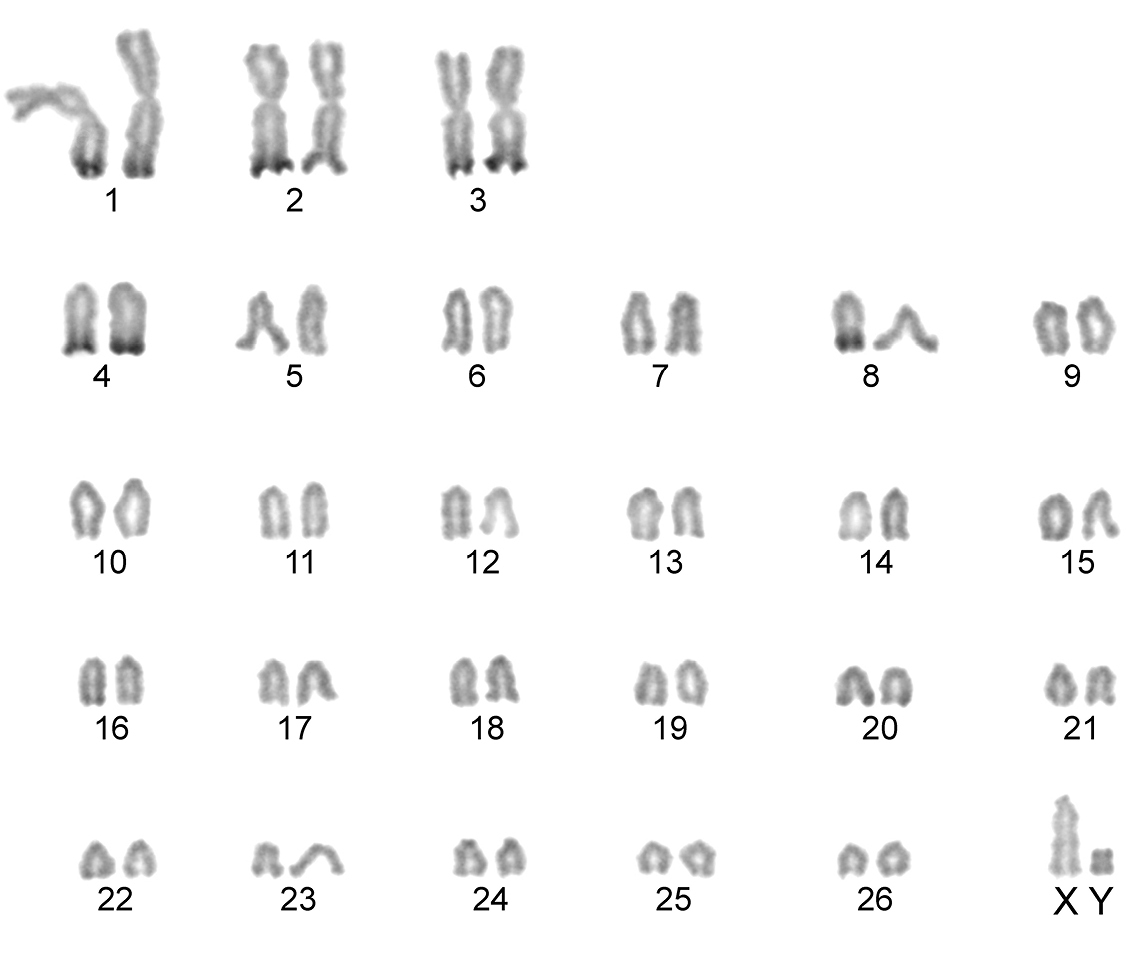
The karyotype contained 54 chromosomes (Fig. 2) including three large metacentric autosomal pairs (no. 1–3) and 23 acrocentric autosomal pairs of decreasing size (nos. 4–26; NFa=58). Tiny short arms were observed in most of the acrocentric autosomes. The X chromosome was a large acrocentric with a distinct short arm, whereas the Y chromosome was metacentric and the smallest element of the complement (NF=61). All the autosomes and both the sex chromosomes were reliably identified on the basis of their unique G-banding patterns (Fig. 3). The C-banded karyotype of Konya wild sheep is illustrated in Fig. 4. All the autosomes possessed distinct and large centromeric C-positive bands which usually extended in the neighbouring pericentromeric areas. The X chromosome had a pericentromeric C-positive band, and the Y chromosome was entirely heterochromatic. Silver-nitrate staining revealed that NORs were localized in the distal position on the long arms of three large metacentric (no. 1–3) and two acrocentric (nos. 4 and 8) autosomes (Fig. 5). All the NORs were observed in both the homologous chromosomes, except those of the pair no. 8 in which a heterozygous condition with the signal present in only one homologue was recorded in four specimens.
Conventionally stained karyotype of the Konya wild sheep with 2n=54. There are three pairs of large metacentric autosomes and 23 pairs of acrocentric autosomes. The X chromosome is acrocentric and the Y chromosome metacentric.
G-banded karyotype of the Konya wild sheep. All the chromosomes in the complement can be identified and differentiated according to their G-banding pattern.
C-banded karyotype of the Konya wild sheep. Distinct pericentromeric dark bands are apparent in all the autosomes and the X chromosome. The Y chromosome is stained entirely positively.
Silver-stained karyotype and the distribution of NORs in the Konya wild sheep. The NORs are localized in telomeric areas of the long arms of the three metacentric autosomal pairs (nos. 1, 2, 3) and two acrocentric autosomal pairs (nos. 4, 8). The heterozygous condition of the NORs in the acrocentric pair no. 8 was found consistently in various cells and different examined individuals.
The obtained karyotypic data confirm possible close
relationships between the Konya wild sheep and domestic sheep. The
diploid number of chromosomes and their morphology expressed in the
number of chromosomal arms and the proportion of the metacentric and
acrocentric autosomes are the same in both complements. The detailed
structure of banded chromosomes of the Konya wild sheep appears quite
similar in comparison with the standard chromosomal complement of the
domestic sheep (cf.
A considerable degree of similarity can also be
demonstrated by comparison of our results with previously reported
differentially stained chromosomes of various wild sheep taxa (
We are obliged to prof. J. Rubeš for his kind advice. This research was supported by the Turkey Nature Protection Head Office of National Parks, and the Ministry of Environment and Forests of Turkey. J.Z. was supported by the Czech Ministry of Education (LC 06073). We are obliged to anonymous reviewers and particularly to the editor for useful comments on the earlier versions of the manuscript.