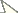(C) 2011 Rodolpho Santos Telles Menezes. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Previous cytogenetic analyses in Trypoxylon Latreille, 1796 have been basically restricted to C-banding. In the present study, base-specific CMA3 and DAPI fluorochrome staining were used to characterize the constitutive heterochromatin in three Trypoxylon species. The heterochromatin was GC-rich in all the species studied; however, in Trypoxylon nitidum F. Smith, 1856the molecular composition of the heterochromatinwasdifferent among chromosome pairs. Conversely, the euchromatin was AT-rich in the three species. These results suggest high conservatism in the euchromatic regions as opposed to the heterochromatic regions that have a high rate of changes. In this study, we report the karyotype of Trypoxylon rugifrons F. Smith, 1873which has the lowest chromosome number in the genus and other characteristics of the likely ancestral Trypoxylon karyotype.
Comparative cytogenetics, heterochromatin, CMA3/DAPI
The Hymenoptera
(Bees, wasps and ants) have received a remarkable amount of attention
due to their amazing diversity of species, life histories, social
behaviors, and key role in very diverse ecosystems. Their importance
arises from their role as pollinators and biocontrol agents, their
damage to agriculture and forestry, and their use as model organisms
for the study of genetics and evolution (
Trypoxylon
Latreille, 1796 is a genus of solitary mud-dauber spider-hunting wasps
that construct tubular nests entirely of mud or in preexisting tubular
cavities. The genus comprises 660 species divided among two subgenera, Trypoxylon s.str. with a cosmopolitan distribution and Trypargilum Richards, 1934 restricted to the Western Hemisphere (
Chromosome number and other karyological features are
good sources of evidence for the systematics and species level taxonomy
of several Hymenoptera groups. A marked chromosomal variability, even within species, has been reported for several aculeate Hymenoptera (
In Trypoxylon, chromosome numbers range from 2n=18 to 2n=34 (
Previous cytogenetic analyses in Trypoxylon have been focused on C-banding, which by itself may not be sufficiently informative for a reliable heterochromatin description and comparative analysis of different species. Therefore, other techniques for molecular characterization can be very useful for this purpose. To improve qualitatively the data available so far, we applied in combination C-banding and base-specific fluorochrome staining to the chromosomes of three Trypoxylon species from the Atlantic rainforest in southern Bahia, in the Brazilian Northeast.
Material and methodsLarvae of Trypoxylon (Trypargilum) nitidum (eight ♀ and three ♂), Trypoxylon (Trypargilum) lactitarse Saussure, 1867 (three ♀ and three ♂) and Trypoxylon (Trypoxylon) rugifrons F. Smith, 1873 (three ♀ and one ♂) were collected in the field directly in the wasp nests for cytogenetic analyses.
Specimens of Trypoxylon nitidum and Trypoxylon lactitarse were captured using trap-nests made of bamboo shoots sectioned below each node with 1 cm diameter or tubes of cardboard with one end closed with the same material and 0.7 cm diameter. The trap-nests were set up in Camacan (15°25'S, 39°29'W) and Ilhéus (14°47'S, 39°12'W), in the state of Bahia and were inspected twice a week. The traps containing complete nests were closed and taken to the Laboratório de Citogenética at the Universidade Estadual de Santa Cruz for the collections of specimens in the prepupal stage. Specimens of Trypoxylon rugifrons were captured from naturally occurring nests in Ilhéus.
At least two specimens per nest were kept at 28°C in a biochemical oxygen demand (BOD) incubator and daily monitored until adult emergence, at which time they were identified. Voucher specimens were deposited at the Entomological Collection at the Universidade Estadual de Santa Cruz.
Slides for cytogenetic analysis were prepared from cerebral ganglia of prepupae following
At least 10 metaphases per specimen were observed using an epifluorescence microscope DMRA2 (Leica) and images were captured using the IM50 Leica software (Leica Microsystems Imaging Solutions Ltd, Cambridge, UK). Giemsa-stained chromosomes were photographed using an Olympus BX51 microscope equipped with an Olympus C-7070 digital camera. Digital images and figure mounting were prepared using Adobe Photoshop CS3 Extended version 10.0.
Chromosomes were described according to Imai’s terminology (
Specimens of Trypoxylon nitidum from Camacan had 2n=30 and n=15 chromosomes and the diploid karyotype formula was 2K=2MCC+10AM+18A (Figs 1A and 1B). Fluorochrome staining (Fig. 1B) revealed that pairs 1 to 5 (AM chromosomes) showed predominantly compacted heterochromatin (equally CMA3/DAPI stained) with balanced GC and AT composition over the long arm, except for the 1st pair, which showed a terminal euchromatic CMA3+/DAPI- (GC-rich) block. The heterochromatin of the acrocentric (6th to 14th pairs) and metacentric (15th pair) chromosomes showed a CMA3+/DAPI- pattern (Fig. 1B) whereas the remaining euchromatin in all chromosomes was CMA3-/DAPI+ (AT-rich). Karyotypes also showed size heteromorphism on the 1st and 5th pairs (Fig. 1B).
Trypoxylon (Trypargilum) nitidum. A Female karyotype (2n=30), standard staining B CMA3/DAPI staining of female karyotype (2n=30). Bar=10µm.
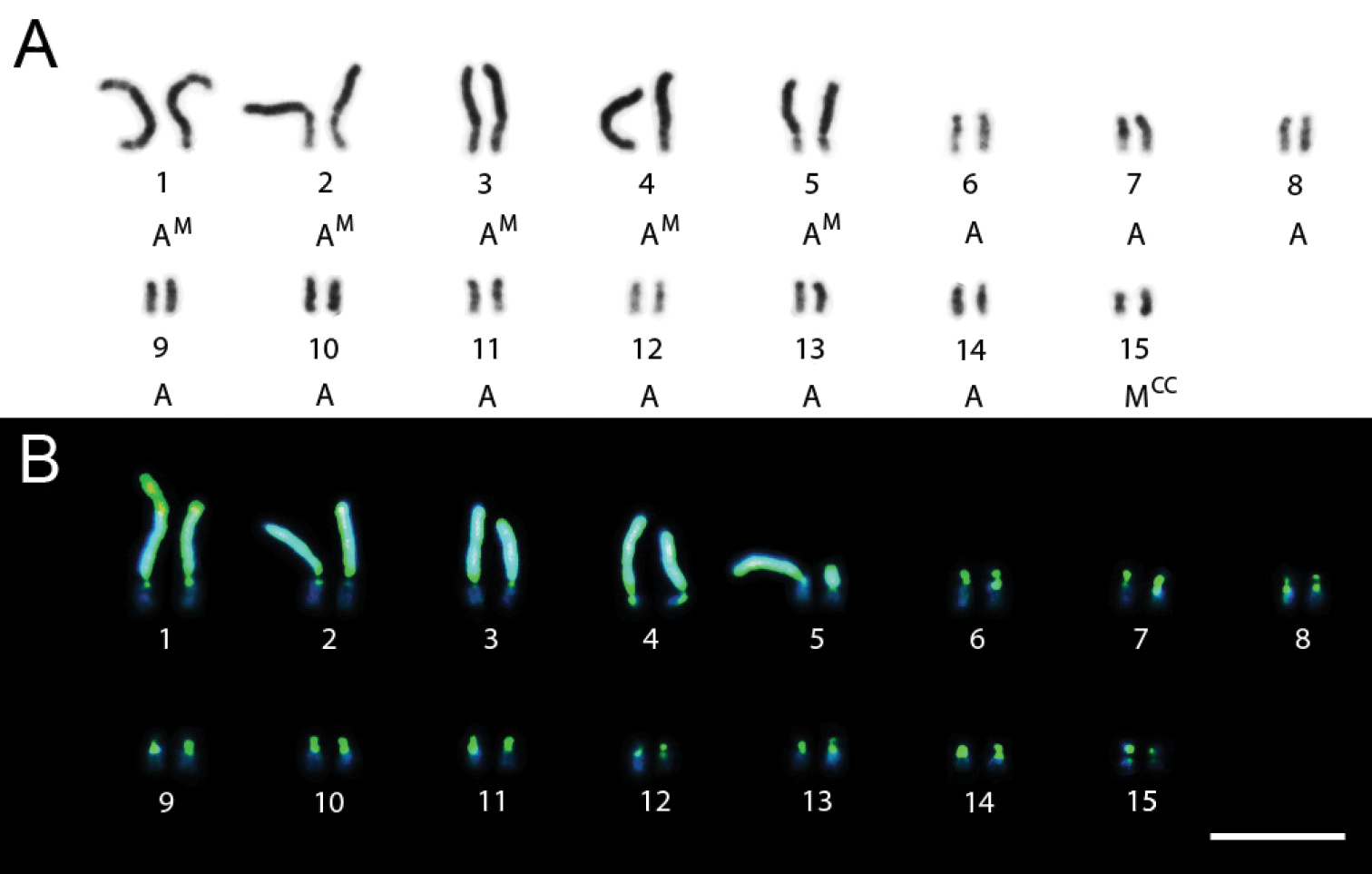
Specimens of Trypoxylon nitidum collected in Ilhéus showed chromosome numbers 2n=29 and n=15 or n=13. The fluorochrome staining pattern also differed in these karyotypes. The 4th pair showed an interstitial CMA3+/DAPI- block near the centromeric region of one of the arms, chromosome pair 15 (Fig. 2B) had the long arm with a CMA3+/DAPI- pattern and chromosome pairs 11 and 13 (Fig. 2A) showed short arms with a CMA3+/DAPI- pattern and long arms with a CMA3-/DAPI+ pattern.
Trypoxylon (Trypargilum) nitidum. A Male karyotype (n=13) with CMA3/DAPI banding B CMA3/DAPI staining of female karyotype (2n=29). Bar=10µm.
Trypoxylon lactitarse had 2n=30 and n=15 chromosomes and the karyotype formula 2K = 2MCC +28AMC (Figs 3A and 3B). The 4th pair was heteromorphic with variation in the length of the long arm (Fig. 3B). The heterochromatin in all chromosomes showed a CMA3+/DAPI- pattern, whereas the euchromatin showed a CMA3-/DAPI+ pattern (Fig. 3B).
Trypoxylon (Trypargilum) lactitarse. A Male karyotype (n=15) with standard staining B CMA3/DAPI staining of female karyotype (2n=30). Bar=10µm.
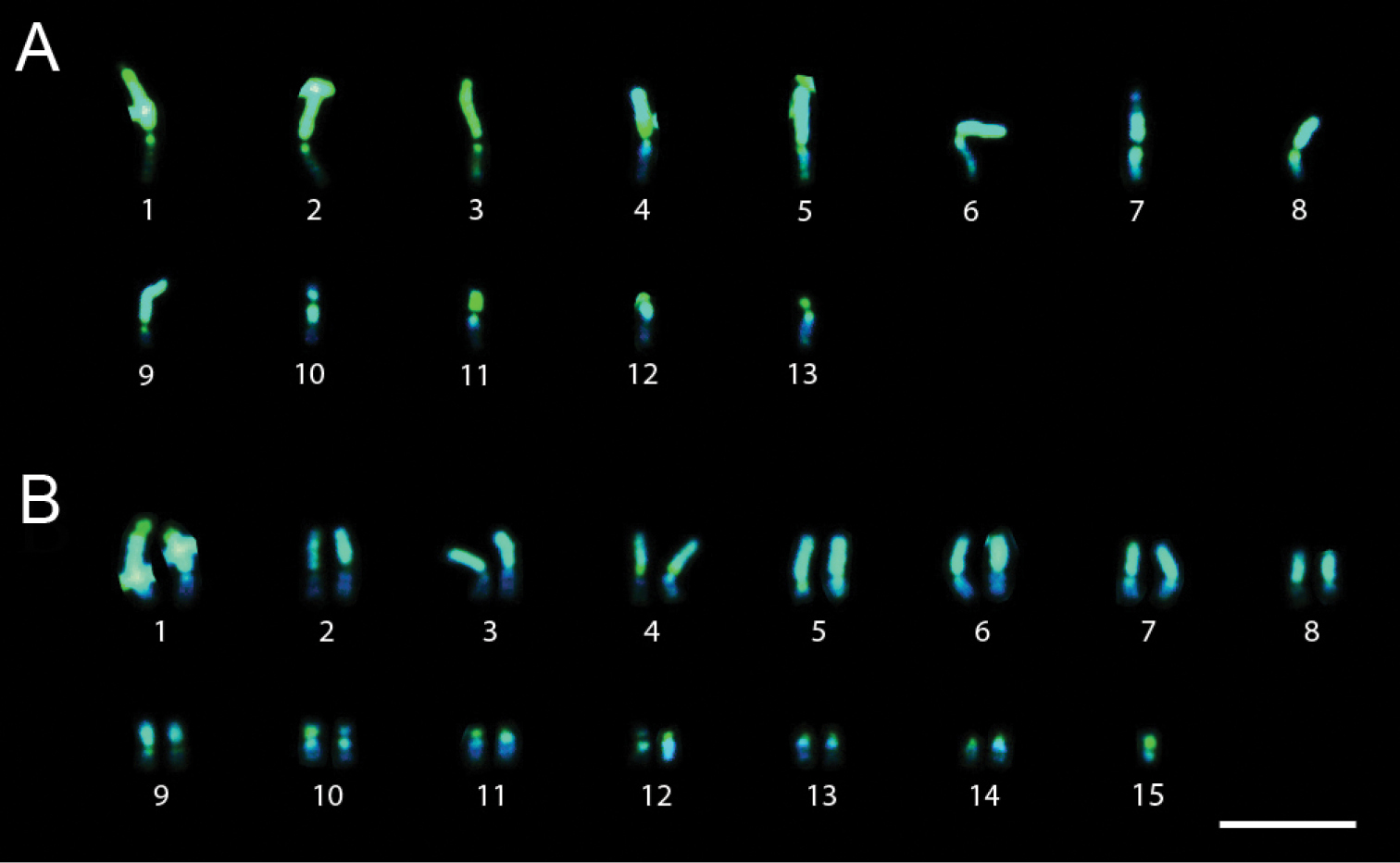
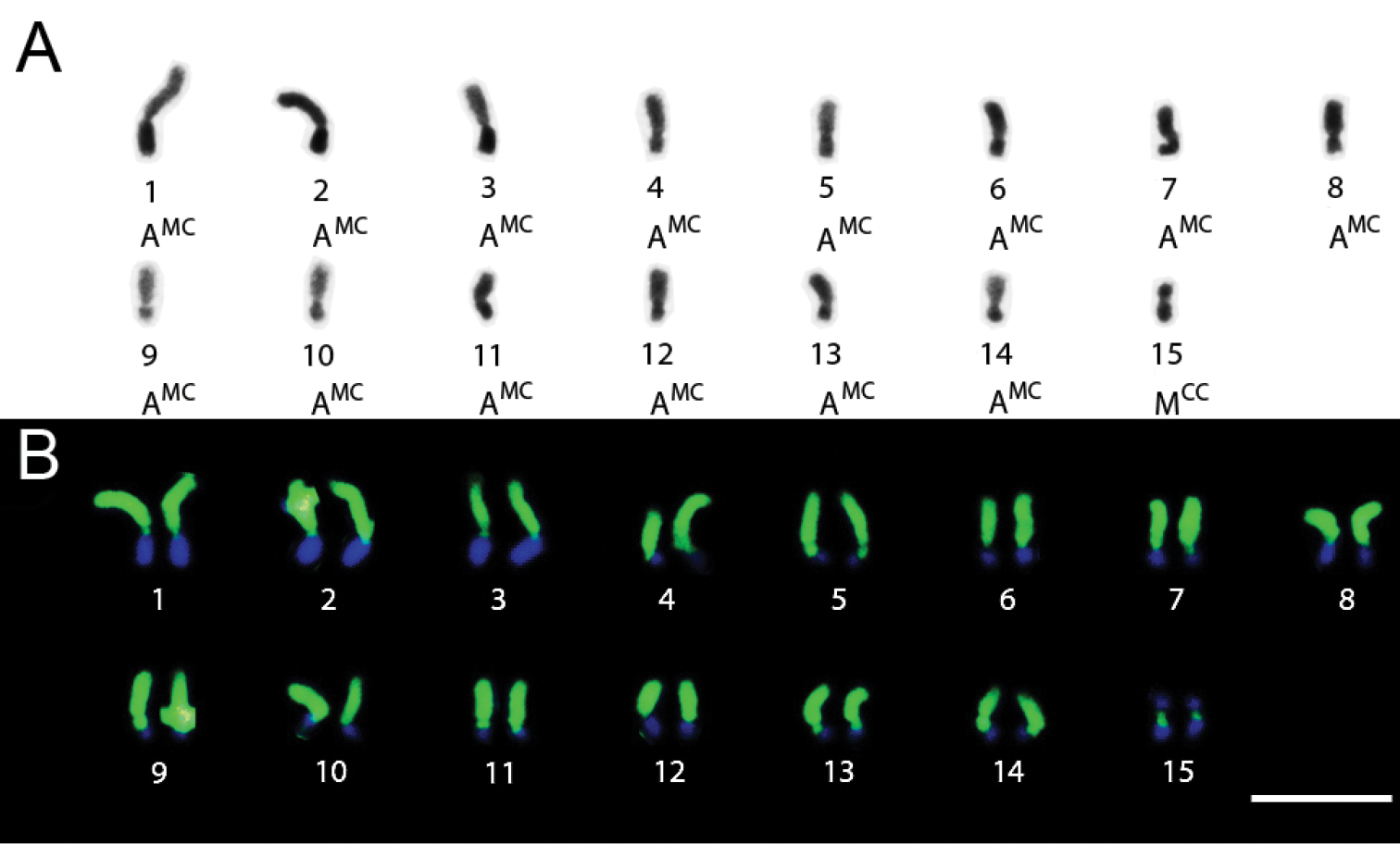
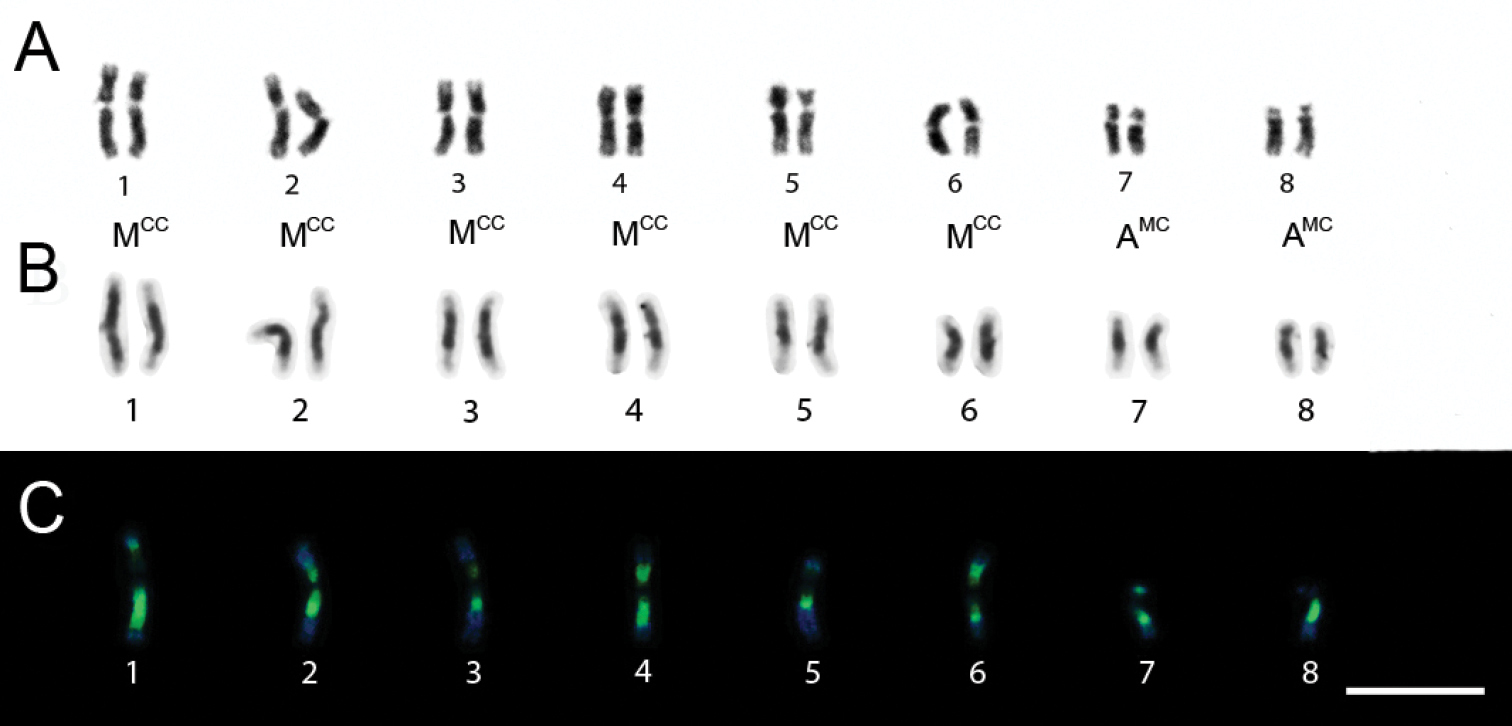
Trypoxylon rugifrons had 2n=16 and n=8 chromosomes and the observed karyotype formula was 2K=12MCC+4AMC (Figs 4A and 4B). The heterochromatin in all chromosomes was CMA3+/DAPI- and the euchromatin showed a CMA3-/DAPI+ pattern (Fig. 4C).
Trypoxylon (Trypoxylon) rugifrons. A Female karyotype (2n=16) with standard staining B C-banding patterns in female karyotype (2n=16) C CMA3/ DAPI staining of male karyotype (n=8). Bar=10µm.
A large variation in chromosome number has been reported for several Hymenoptera groups (
Regarding chromosome morphology, Trypoxylon rugifrons
showed predominantly metacentric and submetacentric chromosomes (pairs 1
to 6) and heterochromatin concentrated in the pericentromeric regions.
These chromosomes are larger than those of the pairs 7 and 8.
The predominance of highly heterochromatic pseudoacrocentric chromosomes in most species within the genus Trypoxylon and the reduced chromosome number and chromosome morphology showed by Trypoxylon rugifrons lend support to Gomes et al.´s (1997) hypothesis. Trypoxylon rugifrons with n = 8 could show features similar to those of the putative ancestral karyotype.
The specimens of Trypoxylon nitidum
from Ilhéus with 2n=29 also had a small metacentric chromosome (15th
pair) with a GC-rich arm whose homologue was not present. These results
have also revealed band similarities between the GC-rich arm and the
terminal region of the 1st pair, which is also GC-rich (Fig. 2B).
This evidence leads us to infer that a fusion between the 15th pair and
the terminal region of one of the chromosomes of pair 1 could be
involved in this numeric variation. This could also explain the
heteromorphism of a CMA3+/DAPI- block present in one of the homologues
of the 1st pair (Fig. 1B).
The heteromorphism found in the 5th pair in Trypoxylon nitidum and in the 4th pair in Trypoxylon lactitarse
could be explained by a deletion or, alternatively, by a tandem
growth of the heterochromatic blocks in the long arms. A previous study
by
Several studies have demonstrated a correspondence between CMA3+ bands and rDNA sites in species such as Scaptotrigona xanthotricha Moure, 1950 and Melipona Illiger, 1806 bees (
The three species studied herein are similar in their
heterochromatin composition. However, the heterochromatin of the
pseudoacrocentric chromosomes in Trypoxylon nitidum
has a balanced GC and AT composition in contrast with the acrocentric
and metacentric chromosomes, which heterochromatin is GC-rich. This
result indicates a difference in the heterochromatin composition in Trypoxylon nitidum. Intraspecific variation in heterochromatin base composition has already been detected in other studies. For example,
The euchromatin was AT-rich in the three species studied, which agrees with
We thank Sérvio Túlio Pires Amarante for species identification; Carter R. Miller and two anonymous reviewers for their valuable comments on a previous version of this manuscript. This study was supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior).