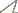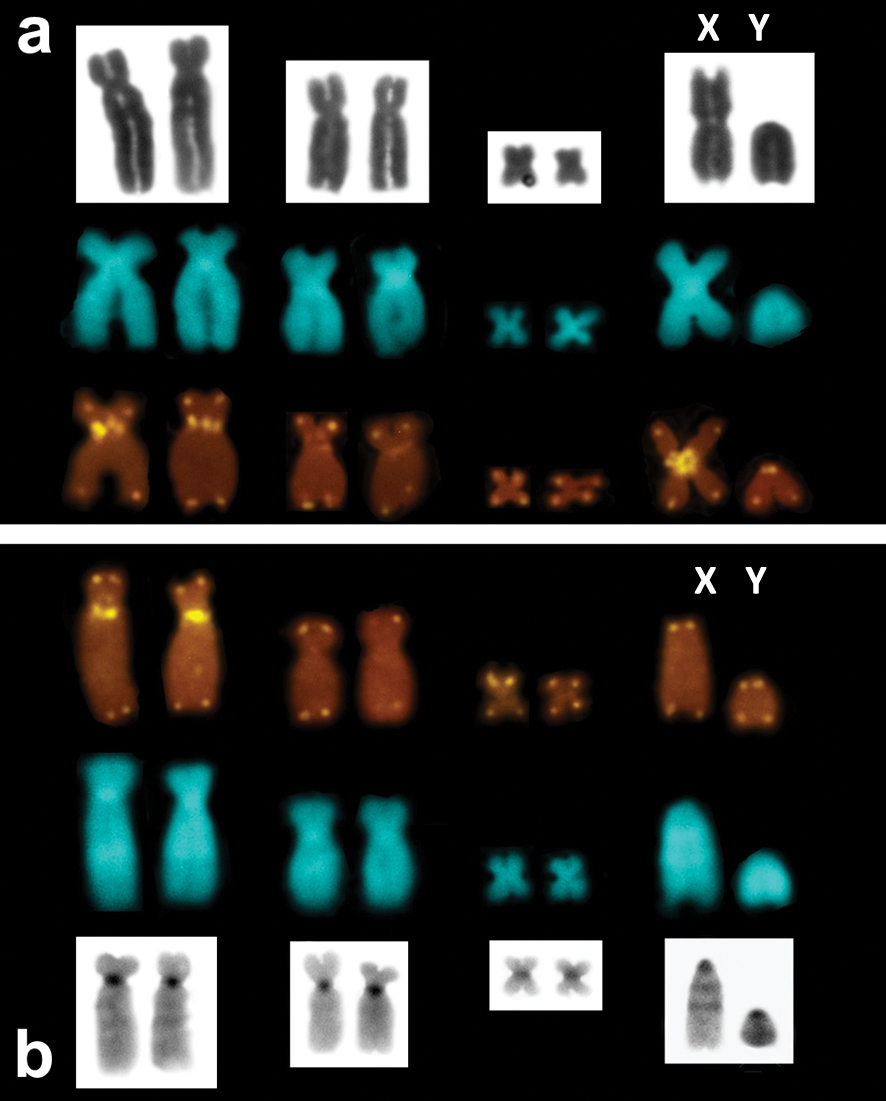(C) 2011 Ekaterina Gornung. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
The Microtus (Terricola) savii s. l. complex is a group of five species/subspecies of the Italian pine voles, which diverged at different times either with or without chromosomal differentiation. The evidence of chromosomal diversification has so far concerned the shape of the sex chromosomes, especially the X chromosome. Three taxa of the group, Microtus savii savii, Microtus savii nebrodensis, and Microtus savii tolfetanus have identical karyotypes with metacentric X chromosomes. The X chromosomes of Microtus brachycercus and Microtus brachycercus niethammericus are, respectively, subtelocentric and acrocentric in shape. The Microtus savii complex has been long an object of conventional karyological studies, but comparative molecular cytogenetic data were completely missing. Therefore, we conducted a comparative chromosomal mapping of rRNA genes (rDNA) and telomeric repeats in three of the five taxa of the group: Microtus savii savii, Microtus savii nebrodensis, and Microtus brachycercus niethammericus, each of which belongs to a distinct mitochondrial clade.The survey revealed that differentiation of the clades was accompanied by remarkable changes with regard to the number and locations of the rDNA sites. Thus, Microtus savii savii and Microtus savii nebrodensis have especially high numbers of rDNA sites, which are located in the centromeric regions of, correspondingly, 18 and 13 chromosome pairs, whereas Microtus brachycercus niethammericus shows variable (8–10) and heteromorphic rDNA sites on both centromeric and telomeric regions. Interstitial telomeric sites (ITS), which are believed to indicate possible breakpoints of recurring chromosomal rearrangements, are present on the largest biarmed chromosomes and on the metacentric X chromosomes in Microtus savii savii and Microtus savii nebrodensis. These preliminary results are discussed in the context of recent advances in phylogeny of the group, as well as the rDNA genomic organization and X chromosome rearrangements in the genus Microtus.
Arvicolinae, chromosomal evolution, sex chromosomes, interstitial telomeric sequences (ITS), rDNA, NORs
The Italian endemic pine voles are distributed throughout the Apennine peninsula from the Alps to Sicily (
The taxonomy and general karyological traits of Microtus savii s. l. complex
| Old taxon1 | New taxon2 | 2n, NFa | Sex chromosomes |
|---|---|---|---|
| Microtus savii savii | Microtus savii savii | 54, 58 | X (m), Y (a) |
| Microtus savii nebrodensis | Microtus savii nebrodensis* | - “ - | - “ - |
| Microtus savii tolfetanus | Microtus savii tolfetanus | - “ - | - “ - |
| Microtus brachycercus | Microtus brachycercus | - “ - | X (sm), Y (a) |
| Microtus savii niethammericus | Microtus brachycercus niethammericus | - “ - | X (a), Y (a) |
m – metacentric, sm – submetacentric, a –
small acrocentric, A – large acrocentric; * – assignment of species
status is possible. 1
Karyological studies in the Microtus savii sensu lato
complex revealed the same diploid number (2n=54) and invariable set of
autosomes (NFa=58) in all these taxa, but only three of the five taxa
showed sex chromosomes similar in size and shape. The sex chromosomes
distinctiveness and the evidence of male sterility of hybrids between “brachycercus” and “savii” supporteda specific rank of “brachycercus”, which was first proposed by
We further investigated the intra- and interspecific
chromosomal variation in the Italian pine voles by analysing chromosomal
distribution of rDNA and telomeric sequences. At present, we focused
on three of the five taxa, i.e. most widespread and abundant Microtus savii savii, the Sicilian Microtus savii nebrodensis, and Microtus brachycercus niethammericus. Each of these taxa belongs to one of the three mitochondrial DNA clades identified in the group (
Specimens of Microtus savii savii
(two males and a female) were collected at three sites: Pizzone
(Isernia, Molise), Parco dell’Appia and Passo Corese (Roma, Lazio).
The specimens of Microtus savii nebrodensis (one male and one female) were trapped on the Nebrodi Mountains (Messina, Sicily). The individuals of Microtus brachycercus niethammericus
(two males and one female) were trapped at Farindola (Pescara,
Abruzzi). The animals were handled according to the European Code of
Practice for the housing and care of animals used in scientific
procedures (
Specimens of Microtus savii savii and Microtus savii nebrodensis showed matching 2n=54 karyotypes with medium-size metacentric X and acrocentric Y chromosomes (Fig. 1a). Specimens of Microtus brachycercus niethammericus had a similar karyotype, but an acrocentric X chromosome of the same size as in the two other taxa. The characteristic C- and DAPI-banding patterns of this X chromosome (Fig. 1b, c) distinguished it from acrocentric autosomes.
Representative karyotypes of the Italian pine voles. A conventional Giemsa stained male karyotype of a Savi’s pine vole exemplified by Microtus savii nebrodensis (A)with a large metacentric X chromosome and a small acrocentric Y. C-banded (B) and DAPI stained (C) chromosome complements of Microtus brachycercus niethammericus, which differ from (A) in morphology of the sex chromosomes. The large acrocentric X chromosomes of Microtus brachycercus niethammericus show distinctive prominent bands. Bar = 10 μm.
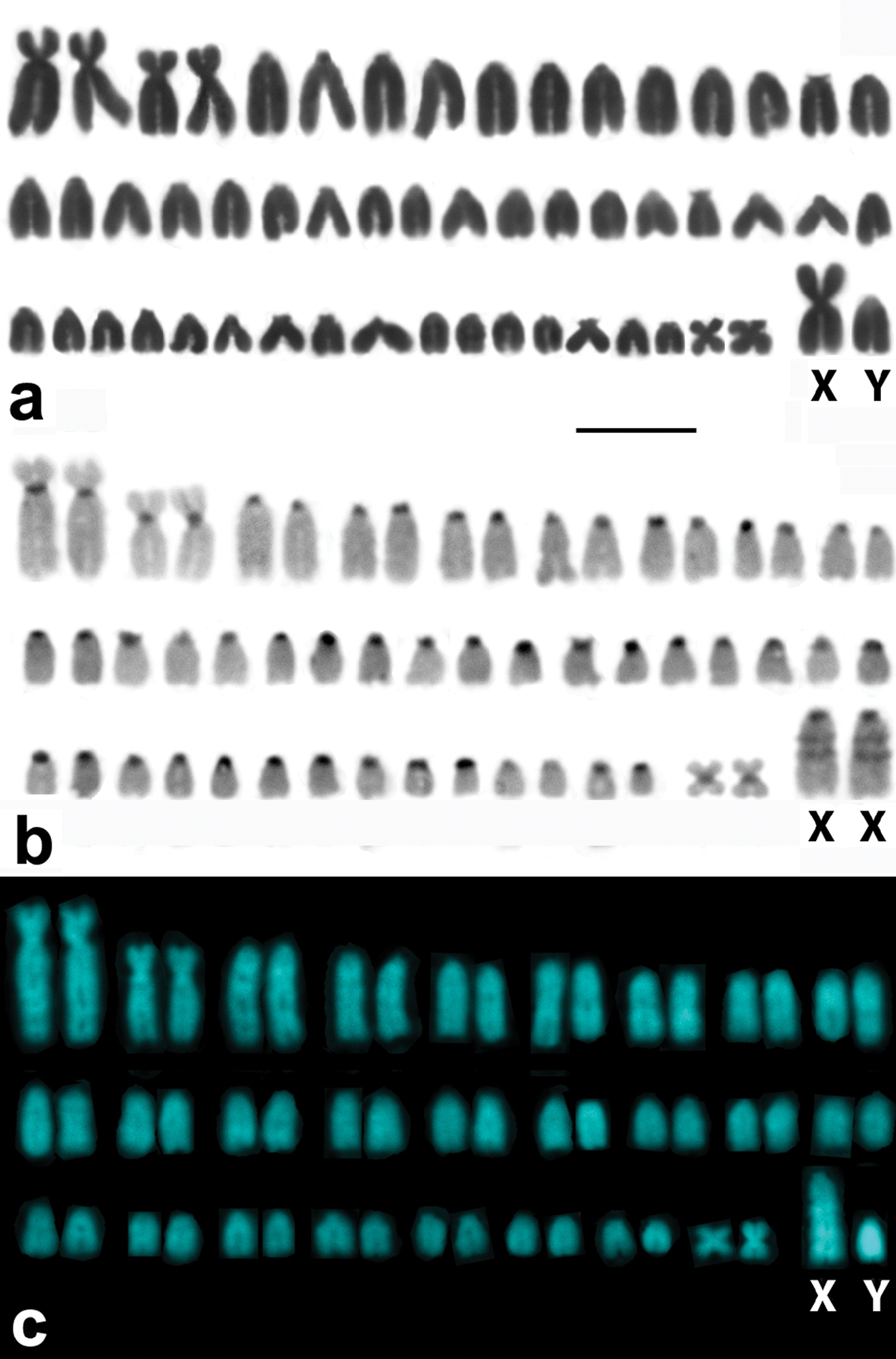
Both the number and locations of the rDNA-FISH signals differed remarkably among the specimens (Fig. 2). The number of signal-bearing chromosomes in metaphase plates of Microtus savii savii was as large as 36 (18 chromosome pairs) (Fig. 2a), while 28 signals were distributed on 13 chromosome pairs in Microtus savii nebrodensis (Fig. 2b). The FISH signals were located at centromeres of acrocentric chromosomes in both Savi’s pine voles and only one pair of medium-sized acrocentric chromosomes of the Sicilian specimens was marked at both chromosome termini (Fig. 2b). The biarmed autosomes and the sex chromosomes lacked rDNA in both taxa (not illustrated). FISH revealed much lower number of rDNA sites per metaphase plate in Microtus brachycercus niethammericus (Fig. 2c). The overall rDNA-FISH pattern remained constant in different metaphase cells of each individual of Microtus brachycercus niethammericus, but differed slightly among presently studied individuals (8, 9 and 10 signals per cell). In this species, hybridization signals were constantly present on two distinct pairs of homologues, a pair of medium-sized acrocentric chromosomes and the smallest pair of metacentric chromosomes, whereas the remaining 4–6 FISH signals were detected on a small set of apparently non-homologous chromosomes. This pattern persisted under various hybridization conditions.
Partial karyotypes composed of rDNA-bearing chromosomes and the sex chromosomes ofMicrotus savii savii (A), M. s. nebrodensis (B), and Microtus brachycercus niethammericus (C). The hybridization signals mark centromeric regions of all NOR-bearing chromosomes in Microtus savii subspecies (A, B) and, additionally, a telomeric region of a single chromosome in (B) (upper row). In (C), the largest among three individuals of Microtus brachycercus niethammericus set of rDNA-bearing chromosomes composed of two constantly marked chromosome pairs (signed by asterisks), one of which represent the smallest biarmed chromosomes, as well as chromosomes with variable rDNA sites, of which two chromosomes are in an apparently heterozygous state. The sex chromosomes lack rDNA-FISH signals in either subspecies.
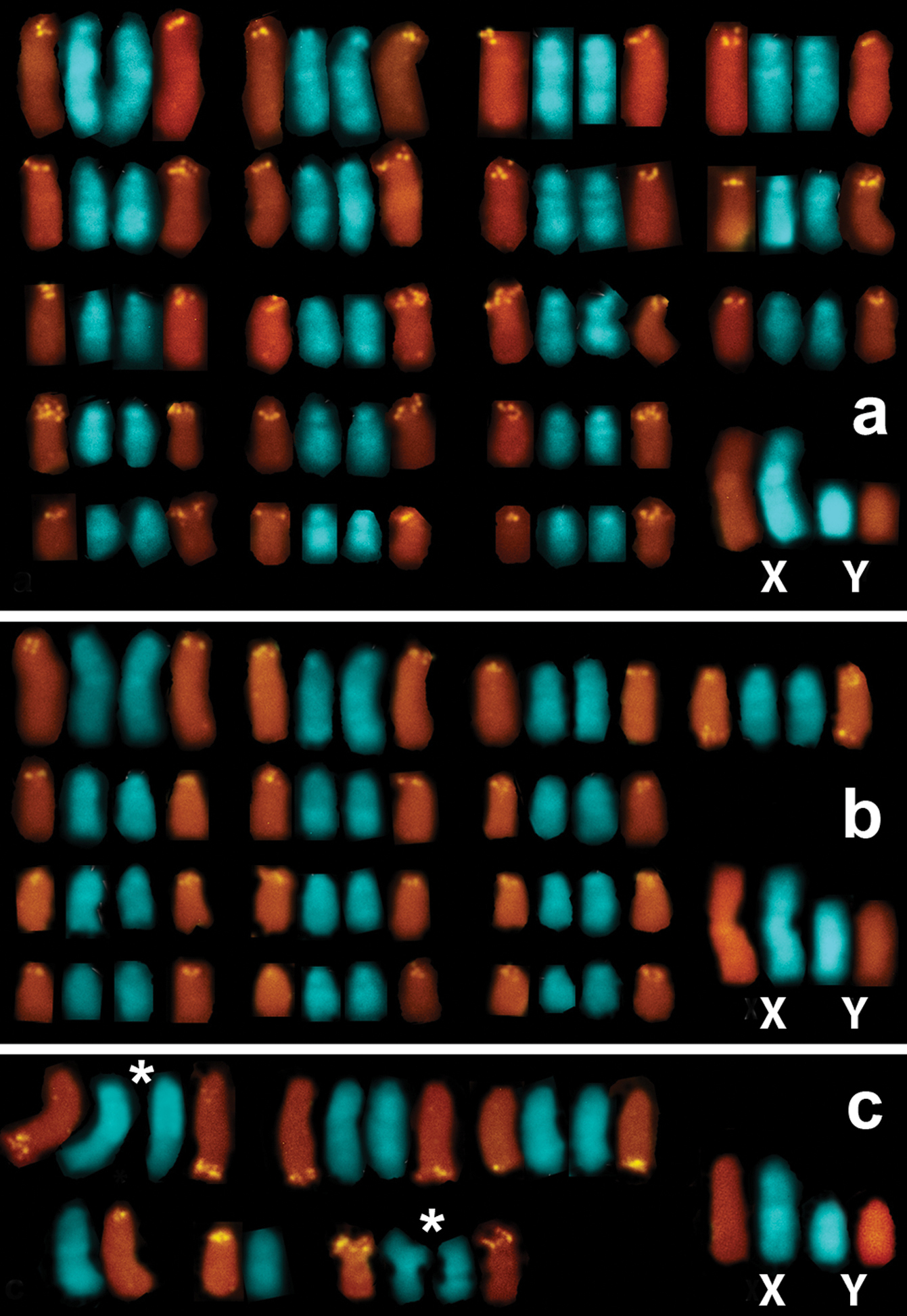
FISH with the telomeric probe followed by high-stringency post-hybridization washes (PHW) showed an ordinary, all-telomeric, pattern in all the specimens studied. Nonetheless, by decreasing the stringency of PHW we revealed telomeric-like sequences on some chromosomes. Thus, prominent ITS signals were present in the centromeric regions of the metacentric X and the largest biarmed (submetacentric) chromosomes of Microtus savii savii and Microtus savii nebrodensis (Fig. 3a). Other chromosomes including two of the three pars of biarmed autosomes (submetacentric and tiny metacentric) and the Y chromosome did not show telomeric signals even under low-stringency conditions. In Microtus brachycercus niethammericus, FISH detected the same ITS pattern on the largest biarmed chromosomes, but the acrocentric X chromosome lacked any interstitial signal (Fig. 3b).
Telomeric FISH signals on thebiarmed and the sex chromosomes of Microtus savii, exemplifiedby Microtus savii savii (A) andMicrotus brachycercus niethammericus (B). Respective chromosome pairs are shown after DAPI counterstaining (central row in A, B), Giemsa staining (upper row in A) and after C-banding (lower row in B). ITS are present on the largest biarmed chromosomes and on the metacentric X chromosome.
The study revealed a considerable variation in the number and chromosomal distribution of rDNA sites at both intraspecific (between Microtus savii savii and Microtus savii nebrodensis)and interspecific (between the two Savi’s pine voles and Microtus brachycercus niethammericus) levels.
The two Savi’s pine voles, Microtus savii savii and Microtus savii nebrodensis, separated in middle-early Pleistocene (0.6–1.0 MYA) (
The rDNA pattern of Microtus brachycercus niethammericus
differs peculiarly from the ones of the congeneric species. The mean
number of rDNA sites is markedly lower. The sites appear variable in
number and size and are located in both centromeric and telomeric
regions of a small set of chromosomes. This evidence is in accord with
the genetic divergence of “brachycercus” clade separated from Microtus savii savii in middle Pleistocene (0.3–0.5 MYA) (
The number and chromosomal locations of NORs (nucleolar
organizer regions, rDNA sites) have been comprehensively studied in
various species of the genus Microtus by conventional silver staining technique (
The increase in the number of NORs in the evolution of
different groups of species, so-called rDNA dispersion, is well
documented. Reciprocal translocations at the level of C bands are
supposed to be the basic underlying mechanism of this event (
Like in all species of Microtus thus studied, except Microtus kirgisorum (
Presently, we show that while interstitial
telomeric-like sequences are marking the largest pair of biarmed
chromosomes in either species, they are also present in the centromeric
region of the metacentric X chromosome of Microtus savii savii and Microtus savii nebrodensis, whereas absent in the heterochromatic regions of the acrocentric X chromosome of Microtus brachycercus niethammericus. We hypothesize that according to the basal position of Microtus savii nebrodensis in the phylogenetic reconstruction (
To date, the evidence of chromosomal diversification in the Microtus savii s.l. complex concerned only the shape of the sex chromosomes, particularly the X chromosome. Presently, we can add several details to this evidence and conclude that significant changes of rDNA genomic organization accompanied the genetic differentiation of the Italian pine voles.
The authors are grateful to Filippo Testagrossa and Antonio Spinnato (Ente Parco Naturale Regionale dei Nebrodi), who provided a permission to collect voles and helped in the fieldwork. This study was supported by funds of “Ateneo Federato della Scienza e della Tecnologia” to R.C. (AST 2008, pr. C26F08W5R2). A.M.R.B. received postdoctoral fellowship from the National Counsel of Technological and Scientific Development (CNPq, pr.150599/2008–0).