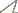(C) 2011 Vladimir E. Gokhman. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Drosophila melanogaster Meigen, 1830 has served as a model insect for over a century. Sequencing of the 11 additional Drosophila Fallen, 1823 species marks substantial progress in comparative genomics of this genus. By comparison, practically nothing is known about the genome size or genome sequences of parasitic wasps of Drosophila. Here, we present the first comparative analysis of genome size and karyotype structures of Drosophila parasitoids of the Leptopilina Förster, 1869 and Ganaspis Förster, 1869 species. The gametic genome size of Ganaspis xanthopoda (Ashmead, 1896) is larger than those of the three Leptopilina species studied. The genome sizes of all parasitic wasps studied here are also larger than those known for all Drosophila species. Surprisingly, genome sizes of these Drosophila parasitoids exceed the average value known for all previously studied Hymenoptera. The haploid chromosome number of both Leptopilina heterotoma (Thomson, 1862) and Leptopilina victoriae Nordlander, 1980 is ten. A chromosomal fusion appears to have produced a distinct karyotype for Leptopilina boulardi (Barbotin, Carton et Keiner-Pillault, 1979)(n = 9), whose genome size is smaller than that of wasps of the Leptopilina heterotoma clade. Like Leptopilina boulardi, the haploid chromosome number for Ganaspis xanthopoda is also nine. Our studies reveal a positive, but non linear, correlation between the genome size and total chromosome length in Drosophila parasitoids. These Drosophila parasitoids differ widely in their host range, and utilize different infection strategies to overcome host defense. Their comparative genomics, in relation to their exceptionally well-characterized hosts, will prove to be valuable for understanding the molecular basis of the host-parasite arms race and how such mechanisms shape the genetic structures of insectcommunities.
Drosophila, Figitidae, parasitoid, genome size, karyotype
Each species has a characteristic genome size and
chromosome number. This information often serves as a starting point for
obtaining whole genome sequence. It is also useful for cytological or
PCR-based genotyping and comparative genomics. Drosophila melanogaster Meigen, 1830 is by far the best-studied insect. Availability of its annotated sequence data (
Many species of Drosophila serve as hosts to parasitic wasps (
Karyotypes of only two parasitic wasps attacking Drosophila spp., namely, Leptopilina heterotoma with n = 10 (
Wasps were cultured on the yw strain of Drosophila melanogaster as described in
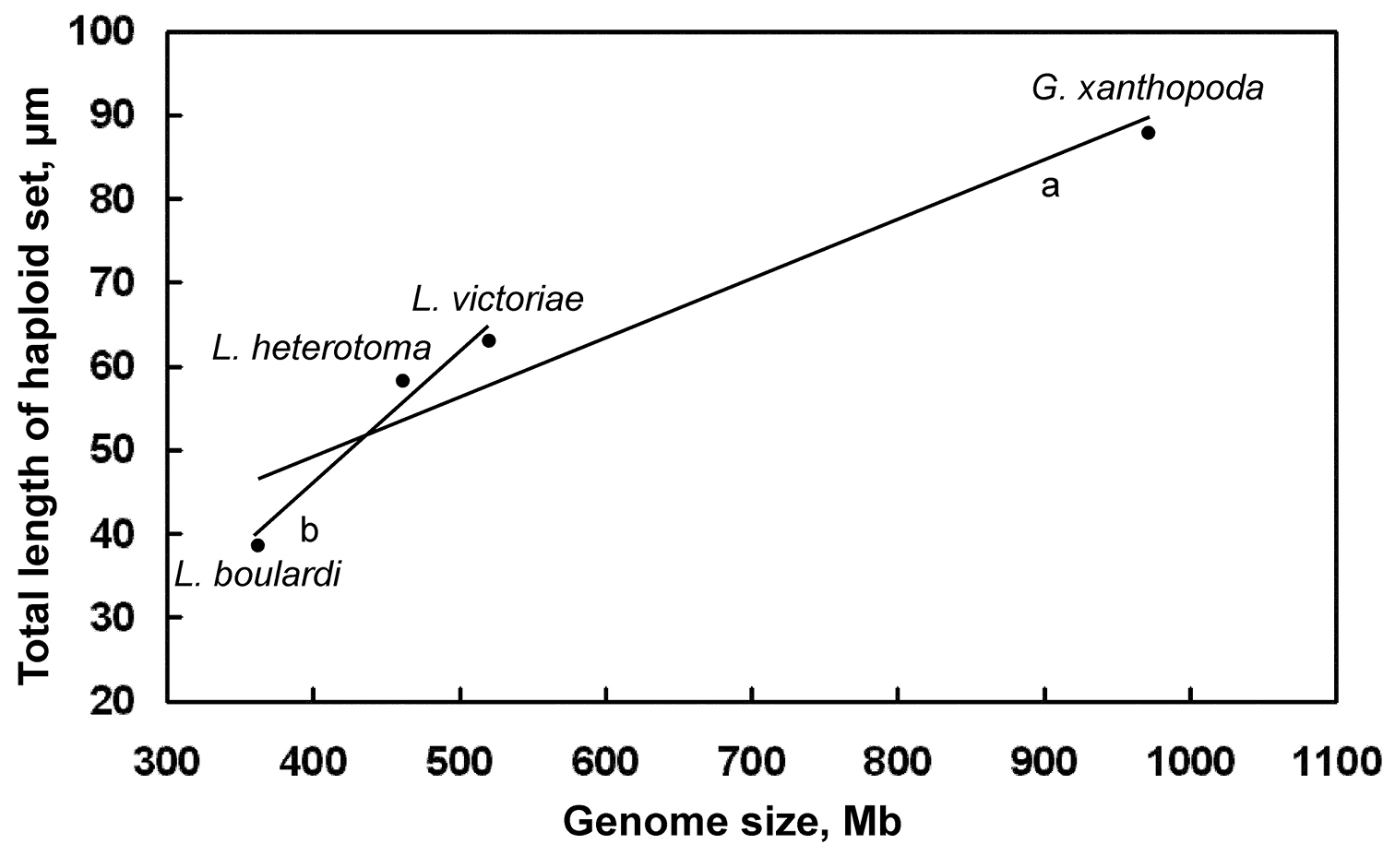
Origins, genome sizes, and gross karyotypic data of Drosophila parasitoids. Genome size of wasp species correlates with total chromosomal length deduced from karyotypic analysis. The total length of the haploid Ganaspis xanthopoda chromosome set differs from both Leptopilina boulardi and Leptopilina heterotoma at p<0.001, and from Leptopilina victoriae at p <0.05; Leptopilina boulardi differs from both Leptopilina heterotoma and Leptopilina victoriae at p<0.001 (T-tests for independent samples).
| Genus, species | Locality, strain | Genome size, mean±SE (Mb), no. specimens studied | Chromosome number, (n) 2n/no. (haploid) diploid specimens studied | Total length of haploid set, mean±SE (μm)/no. metaphases studied | Reference/note |
|---|---|---|---|---|---|
| Ganaspis xanthopoda | New York | 971.5±6.7/4 | (9)/(2) | 87.7±8.3/3 |
|
| Leptopilina boulardi | G486 | 370.0±3.2/5 | (9)18/(1)1 | Not studied |
|
| Leptopilina boulardi | 17 | 362.8±1.7/5 | (9)18/(7)4 | 38.6±3.0/7 |
|
| Leptopilina boulardi | France | 366.0±2.2/5 | Not studied | Not studied |
|
| Leptopilina boulardi | Average | 366.3±2.4/15 | (9)18/(8)5 | 38.6±3.0/7 | Pooled data |
| Leptopilina heterotoma | New York | 461.9±1.9/6 | (10)20/(6)9 | 58.3±2.1/17 |
|
| Leptopilina heterotoma | 14 | 460.0±1.4/5 | (10)20/(3)5 | Not studied |
|
| Leptopilina heterotoma | Average | 460.9±1.7/11 | (10)20/(9)14 | 58.3±2.1/17 | Pooled data |
| Leptopilina victoriae | The Netherlands | 520.2±0.8/5 | (10)/(3) | 63.1±4.5/5 |
|
| Leptopilina (genus) | Average | 424.7±11.0/31 | N/A/(20)19 | 54.4±2.3/29 | Pooled data |
Flow cytometric analysis of genome size, based on nuclei isolated from heads of females of three species of Leptopilina, and Ganaspsis xanthopoda was carried out as described before (
Chromosomal preparations for karyology were obtained from
cerebral ganglia of male and female prepupae of parasitic wasps
according to the technique used by
The results of the study of genome sizes of the Drosophila parasitoids show almost no intraspecific variation, yet greater than 2.5-fold interspecific variation (Fig. 1; Table 1). The gametic genome size of Ganaspis xanthopoda (1 C = 971 Mb) is larger than that of any of the three Leptopilina species (370 Mb < 1C < 520 Mb) studied (Fig. 1). In turn, the genome sizes of all parasitic wasps studied in this paper are also larger than those known for all Drosophila species, which range from 1C = 136.5 to 331.5 Mb (
Cytograms showing relative fluorescence and total propidium iodide-stained nuclei of samples and standards to determine genome size. a relative fluorescence of PI-stained 2C nuclei from one head of a Ganaspis xanthopoda strain NY female co-prepared with 2C and 4C nuclei from one head of a Drosophila virilis female standard (1C = 333 Mb) b–d relative fluorescence and total PI stained nuclei of co-prepared Leptopilina and Drosophila melanogaster (1C = 175 Mb) to determine genome size for Leptopilina boulardi (panel b), Leptopilina heterotoma (panel c), and Leptopilina victoriae (panel d). Genome size is calculated as follows: (mean fluorescence channel number of sample 2C peak/mean fluorescence channel number of 2C standard peak) X 1C DNA content of the standard, with the genome size mean and standard error calculated from repeat co-preparations using different individuals of each species.
Our results provide the first information on genome sizes not only of the family Figitidae, but of the superfamiy Cynipoidea
as a whole. It is intriguing that the genome sizes of all these
parasitoids exceed the average value known for previously studied Hymenoptera, i.e., 360.75 Mb (
Total lengths of haploid chromosome sets of above mentioned species are given in Table 1. The relative lengths and centromere indices of all chromosomes are given in Table 2.
Relative lengths (RL) and centromere indices (CI) of Drosophila
parasitoids. (mean±SE). Strains and numbers of studied metaphase plates
are as in Table 1. Centromere indices are: metacentrics: 37.5-50.0;
submetacentrics: 25.0-37.5; subtelocentrics: 12.5-25.0; acrocentrics:
0-12.5, according to
| Species/ chromosome no. | Ganaspis xanthopoda | Leptopilina boulardi | Leptopilina heterotoma | Leptopilina victoriae | ||||
|---|---|---|---|---|---|---|---|---|
| RL | CI | RL | CI | RL | CI | RL | CI | |
| 1 | 24.17± 0.77 | 37.50± 5.34 | 31.13± 0.81 | 39.97± 2.98 | 14.21± 0.32 | 28.26± 1.42 | 15.49± 0.21 | 39.27± 4.55 |
| 2 | 12.85± 0.25 | 20.50± 1.57 | 13.06± 0.37 | 35.13± 2.79 | 11.89± 0.16 | 30.03± 1.07 | 11.63± 0.20 | 30.46± 4.97 |
| 3 | 11.97± 0.11 | 20.14± 1.28 | 11.45± 0.27 | 29.86± 2.56 | 11.01± 0.11 | 28.58± 1.44 | 11.04± 0.17 | 32.95± 5.05 |
| 4 | 10.59± 0.32 | 19.80± 3.50 | 9.19± 0.17 | 21.03± 2.99 | 10.51± 0.82 | 27.72± 1.16 | 10.48± 0.19 | 31.88± 4.34 |
| 5 | 9.35± 0.61 | 22.25± 3.56 | 8.54± 0.19 | 18.66± 4.06 | 10.02± 0.69 | 28.90± 1.93 | 9.53± 0.13 | 33.69± 5.72 |
| 6 | 8.75± 0.43 | 35.55± 2.49 | 7.32± 0.19 | 17.33± 2.79 | 9.40± 0.11 | 33.17± 1.89 | 9.15± 0.13 | 31.12± 3.84 |
| 7 | 8.39± 0.15 | 15.86± 0.39 | 6.95± 0.15 | 11.86± 3.68 | 8.92± 0.11 | 32.12± 1.96 | 9.02± 0.11 | 34.72± 4.95 |
| 8 | 7.81± 0.42 | 43.48± 1.32 | 6.42± 0.10 | 13.91± 4.84 | 8.48± 0.12 | 30.16± 1.54 | 8.69± 0.08 | 41.12± 2.45 |
| 9 | 6.12± 0.10 | 1.44± 0.73 | 5.94± 0.13 | 8.77± 2.60 | 8.04± 0.11 | 28.70± 1.49 | 8.01± 0.26 | 34.88± 2.57 |
| 10 | - | - | - | - | 7.52± 0.09 | 31.93± 1.88 | 6.96± 0.28 | 36.19± 1.87 |
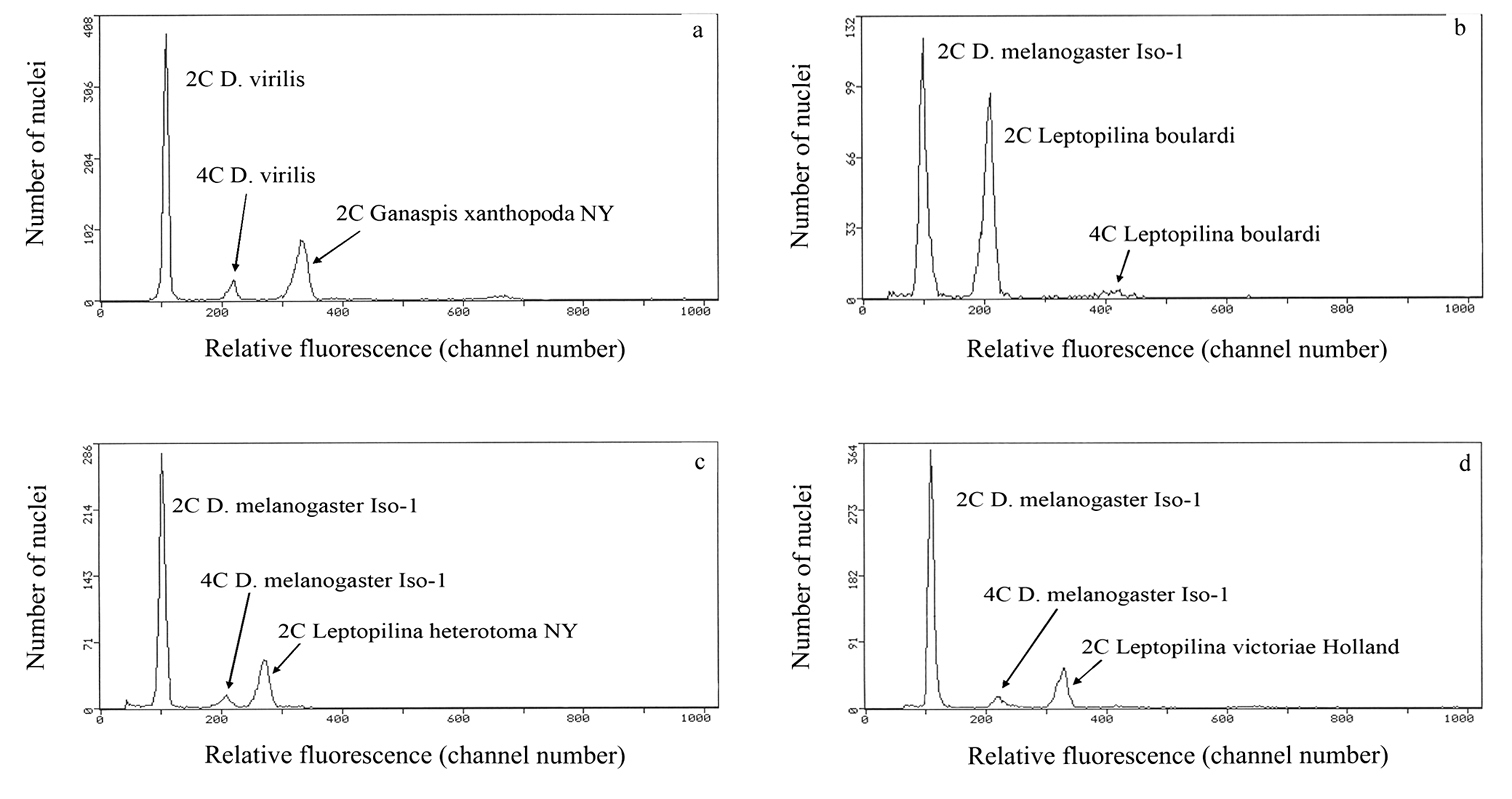
Ganaspis xanthopoda. Nine chromosomes were found in the haploid karyotype of this species (n = 9; Fig. 2a). Chromosomes are long relative to Leptopilina spp. (see Table 1 and below); most of them are of similar size. However, the first meta- or submetacentric chromosome is about twice as long as the remaining ones. Most other chromosomes are subtelocentric, except for the sixth submetacentric, eighth metacentric, and last acrocentric ones.
Karyograms of Drosophila parasitoids. a Ganaspis xanthopoda, haploid set b Leptopilina boulardi (strain 17), haploid set c ditto, diploid set d Leptopilina heterotoma (New York strain), haploid set e ditto, diploid set f Leptopilina victoriae, haploid set. Scale bar 10 μm.
Leptopilina boulardi. As in Ganaspis xanthopoda, n = 9 (and 2n = 18; Figs 2b and c; Fig. 3a). Moreover, the karyotype of Ganaspis xanthopoda is superficially similar to that of Leptopilina boulardi in that the very large first metacentric chromosome is more than twice as long as the second. However, the length of all remaining Leptopilina boulardi chromosomes is roughly half that of the Ganaspis xanthopoda chromosomes. Furthermore, chromosomes of the second and third pairs are submetacentric, those of the fourth, fifth, sixth and eighth pairs are subtelocentric, and chromosomes of the seventh and ninth pairs are acrocentric.
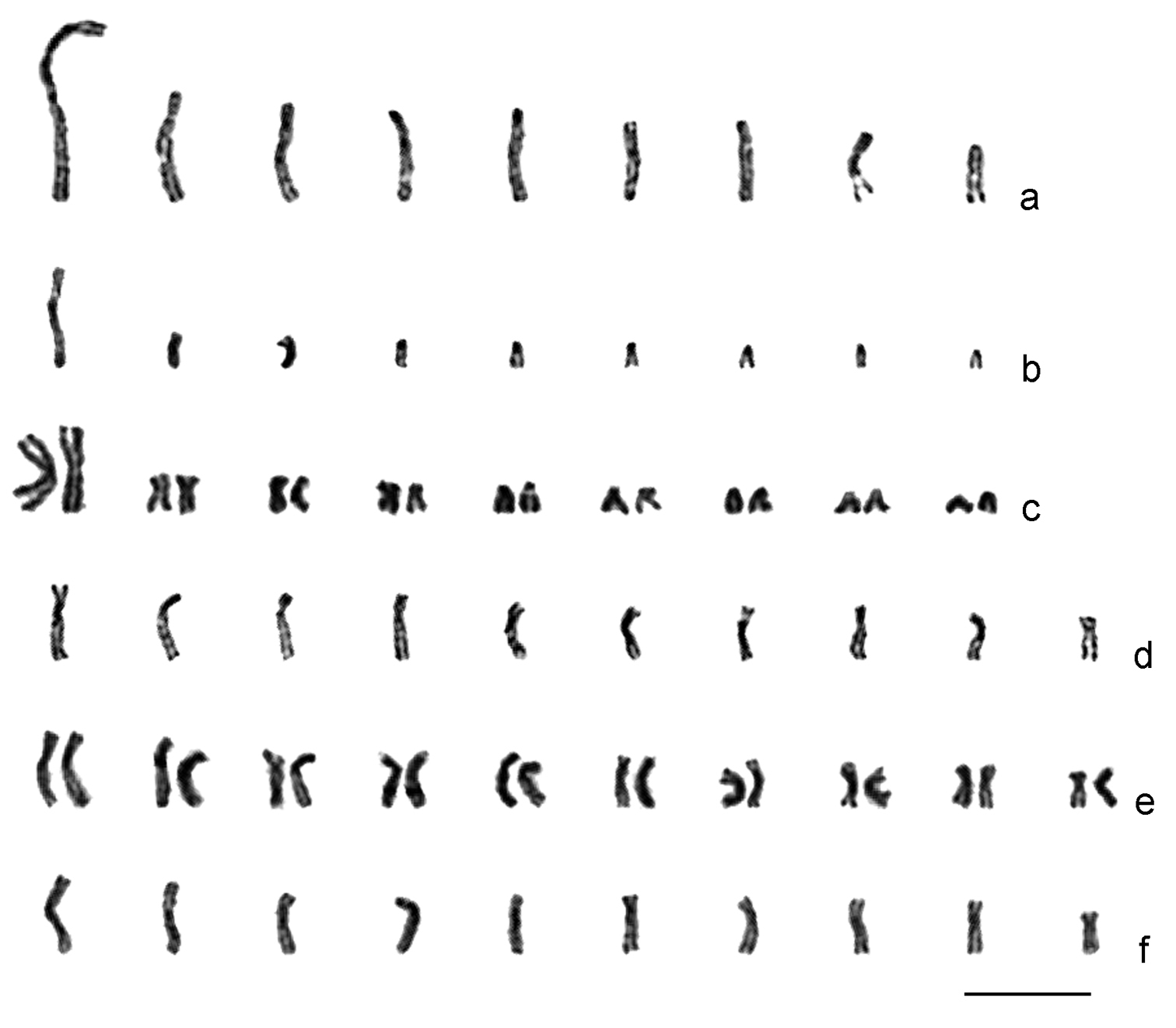
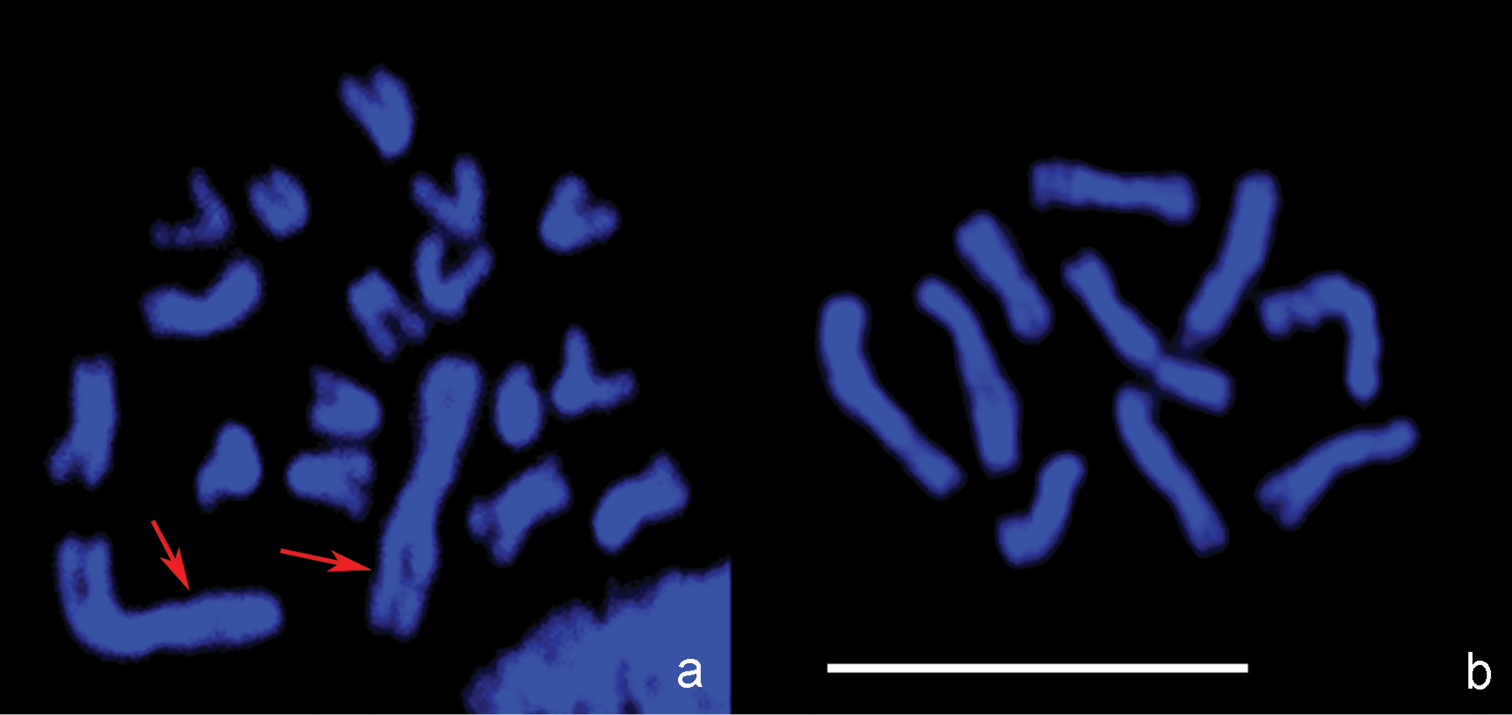
Confocal microscopic images. a Leptopilina boulardi, diploid metaphase plate b- Leptopilina heterotoma, haploid metaphase plate. Arrows point to the pair of large metacentric chromosomes in the karyotype of Leptopilina boulardi that presumably arose via chromosomal fusion in an ancestral chromosome set with n = 10. Scale bar 10 μm.
Leptopilina heterotoma. Consistent with previous observations (
Leptopilina victoriae. This species belongs to the Leptopilina heterotoma clade (
Parasitic wasps make up a significant number of species of all insects (
In a study of genome size of 89 species of bees, wasps, and ants,
Our karyotypic study provides new insights into the genome structure of Drosophila
parasitoids. First, the study demonstrates an obvious positive
correlation between the genome size and total chromosome length in those
parasitic wasps (Table 1; Fig. 4). However, chromosome length in Ganaspis xanthopoda increases relatively slower than might be expected from its larger genome size (Fig. 4). This observation suggests that a significant portion of the bloated Ganaspis
genome is repeat sequence that is highly condensed at metaphase. High
copy number of satellite DNA is associated with genome size variation in
Drosophila species (
Distribution of genome size/chromosome length of Drosophila parasitoids. Mean values are given for each species. Trend lines: a for all species combined b for Leptopilina spp. (i.e. all species excluding Ganaspis xanthopoda).
Second, our study reveals that genome sizes vary independently of the chromosome number in Drosophila parasitoids. This may not be surprising if the large metacentric chromosomes of Leptopilina boulardi (Fig. 2b; Fig. 3a) and Ganaspis xanthopoda (Fig. 2a)
have an independent origin via chromosomal fusions. Parallel
chromosomal fusions are relatively frequent within various lineages of
parasitic Hymenoptera (
Third, the karyotype provides the scaffold number for
future sequencing effort in these insects. When the karyotypic features
of the species studied here are superimposed onto their phylogeny (
The authors are grateful to Andrey P. Mikhailenko (MSU Botanical Garden) for technical assistance. This work was supported by grants from the Russian Foundation for Basic Research (10-04-01521) to V.E.G., and from the U.S. Department of Agriculture (NRI/USDA CSREES 2006-03817 and NIFA 2009-35302-05277); National Institute of Health (NIGMS S06 GM08168, GM056833-10, P50-GM68762, and G12-RR03060); and the Professional Staff Congress of the City University of New York (S.G.), and a National Human Genome Research Institute (NHGRI) subcontract to J.S.J. The authors declare no conflict of interest.