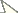(C) 2011 Andrei Sourakov. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
The genus Calisto Hübner, 1823 is the only member of the diverse, global subfamily Satyrinae found in the West Indies, and by far the richest endemic Caribbean butterfly radiation. Calisto species occupy an extremely diverse array of habitats, suggestive of adaptive radiation on the scale of other classic examples such as the Galápagos or Darwin’s finches. However, a reliable species classification is a key requisite before further evolutionary or ecological research. An analysis of 111 DNA ‘barcodes’ (655 bp of the mitochondrial gene COI) from 29 putative Calisto species represented by 31 putative taxa was therefore conducted to elucidate taxonomic relationships among these often highly cryptic and confusing taxa. The sympatric, morphologically and ecologically similar taxa Calisto confusa Lathy, 1899 and Calisto confusa debarriera Clench, 1943 proved to be extremely divergent, and we therefore recognize Calisto debarriera stat. n. as a distinct species, with Calisto neiba Schwartz & Gali, 1984 as a junior synonym syn. n. Species status of certain allopatric, morphologically similar sister species has been confirmed: Calisto hysius (Godart, 1824) (including its subspecies Calisto hysius aleucosticha Correa et Schwartz, 1986, stat. n.), and its former subspecies Calisto batesi Michener, 1943 showed a high degree of divergence (above 6%) and should be considered separate species. Calisto lyceius Bates, 1935/Calisto crypta Gali, 1985/Calisto franciscoi Gali, 1985 complex, also showed a high degree of divergence (above 6%), confirming the species status of these taxa. In contrast, our data suggest that the Calisto grannus Bates, 1939 species complex (including Calisto grannus dilemma González, 1987, Calisto grannus amazona González, 1987, stat. n., Calisto grannus micrommata Schwartz & Gali, 1984, stat. n., Calisto grannus dystacta González, 1987, stat. n., Calisto grannus phoinix González, 1987, stat. n., Calisto grannus sommeri Schwartz & Gali, 1984, stat. n., and Calisto grannus micheneri Clench, 1944, stat. n.) should be treated as a single polytypic species, as genetic divergence among sampled populations representing these taxa is low (and stable morphological apomorphies are absent). A widely-distributed pest of sugar cane, Calisto pulchella Lathy, 1899 showed higher diversification among isolated populations (3.5%) than expected, hence supporting former separation of this species into two taxa (pulchella and darlingtoni Clench, 1943), of which the latter might prove to be a separate species rather than subspecies. The taxonomic revisions presented here result in Calisto now containing 34 species and 17 subspecies. Three species endemic to islands other than Hispaniola appear to be derived lineages of various Hispaniolan clades, indicating ancient dispersal events from Hispaniola to Puerto Rico, Cuba, and Jamaica. Overall, the degree of intrageneric and intraspecific divergence within Calisto suggests a long and continuous diversification period of 4–8 Myr. The maximum divergence within the genus (ca. 13.3%) is almost equivalent to the maximum divergence of Calisto from the distant pronophiline relative Auca Hayward, 1953 from the southern Andes (14.1%) and from the presumed closest relative Eretris Thieme, 1905 (14.4%), suggesting that the genus began to diversify soon after its split from its continental sister taxon. In general, this ‘barcode’ divergence corresponds to the high degree of morphological and ecological variation found among major lineages within the genus.
COI, biogeography, DNA barcoding, islands, intraspecific variation, Lepidoptera , Nymphalidae, Satyrinae, speciation, taxonomy
The genus Calisto Hübner, 1823 is endemic to the West Indies, and, until the present revision, comprised 54 named taxa (
Though Calisto are neither visually spectacular nor economically important (with the exception of Calisto pulchella
Lathy, 1899 , which is a pest of sugar cane), a significant amount
of information is available on the distribution of the more common
species on Hispaniola from the general survey of the island’s
butterflies by
The morphology of immature stages has been utilized extensively in phylogenetic studies of butterflies (e. g.,
Many species of Calisto were described only recently, towards the end of the 20th century (e.g.,
A different issue is presented by the taxa that are
clearly allopatric (and probably remained in isolation for a long time),
but which are so morphologically similar that one must question the
extent of diversification between them. For instance,
We find allopatric similar taxa within other major species groups, such as Calisto chrysaoros Bates, 1935 (names include Calisto galii Schwartz, 1983 and galii choneupsilon Schwartz, 1985) and Calisto lyceius Bates, 1935 (names include Calisto crypta Gali, 1985 and Calisto franciscoi Gali, 1985). In the Calisto confusa Lathy, 1899 complex, the name Calisto confusa debarriera
Clench, 1943 has been attributed to a form with reduced white discal
and extradiscal bands on the underside, which is found throughout the
geographic range of Calisto confusa confusa and is occasionally sympatric, though frequently replaces typical Calisto confusa phenotypes at higher elevations. Calisto montana
Clench, 1943 was described from the same group based on a single very
worn specimen which had an unusual double-pupiled eye-spot on the
underside of its forewing (Fig. 7) – a character found occasionally throughout Calisto. Other taxa within Calisto confusa species complex have also been described, such as Calisto gonzalezi Schwartz, 1988 for which
The above confusion over the recently described taxa is
perhaps partly due to sole reliance of the authors on wing
characteristics combined with distribution data in their approach to
delineating new species, partly due to limited series and quality of
specimens, and partly due to the exercising of the typological approach
in its extreme form, with a disregard for interspecific variation. A
possible solution to the problem is to use a new set of characters such
as molecular sequence data. The technique of ‘DNA barcoding’ is based on
the analysis of short, standardized gene regions; in the case of
animals, this is a 655-bp segment of mitochondrial cytochrome oxidase
subunit I (COI). DNA barcoding potentially provides an efficient method
for species identification as well as for solving species-level
taxonomical problems. Although the DNA barcode region can vary
intraspecifically on a geographic scale as well as within populations
(e. g.,
A total of 110 Calisto specimens representing 31 putative taxa were sampled (Table 1). All specimens were collected in 1994–1999 by the first author. None of the specimens were subjected to any chemical treatment before desiccation. The climate of the regions ensured quick drying of specimens, which were stored at a room temperature (18–25°C) for over 10 years. DNA was extracted from a single leg removed from each specimen. Specimens were mostly unprepared (papered), with the exception of several individuals.
We amplified a 655-bp segment of mitochondrial cytochrome oxidase subunit I, from the COI
barcode region. All polymerase chain reactions and DNA sequencing were
carried out following standard DNA barcoding procedures for Lepidoptera as described previously (
We chose two genera as outgroups: Eretris, which
Sequences were aligned using BioEdit software (
Sequence data were analyzed using Bayesian inference (BI), as implemented in Mr Bayes 3.1.2 (
Calisto species examined in the present study and resulting nomenclatural changes.
| Smith & al. 1994 name | Describer(s) | Status change | Proposed new status |
|---|---|---|---|
| Calisto aleucosticha | Correa & Schwartz, 1986 | stat. n. | Calisto hysius aleucosticha |
| Calisto amazona | González, 1987 | stat. n. | Calisto grannus amazona |
| Calisto arcas | M. Bates, 1939 | ||
| Calisto archebates | (Ménétriés, 1832) (Satyrus) | ||
| Calisto batesi | Michener, 1943 | ||
| Calisto chrysaoros | M. Bates, 1935 | ||
| Calisto confusa | Lathy, 1899 | ||
| Calisto confusa debarriera | Clench, 1943 | stat. n. | Calisto debarriera |
| Calisto crypta | Gali, 1985 | ||
| Calisto dystacta | González, 1987 | stat. n. | Calisto grannus dystacta |
| Calisto eleleus | M. Bates, 1935 | ||
| Calisto franciscoi | Gali, 1985 | ||
| Calisto gonzalezi | Schwartz, 1988 | syn. n. | Calisto debarriera |
| Calisto grannus | M. Bates, 1939 | ||
| Calisto grannus dilemma | González, 1987 | ||
| Calisto herophile | Hübner, [1823] | ||
| Calisto hysius | (Godart, [1824]) (Satyrus) | ||
| Calisto lyceius | M. Bates, 1935 | ||
| Calisto micheneri | Clench, 1944, repl. name | stat. n. | Calisto grannus micheneri |
| Calisto micrommata | Schwartz & Gali, 1984 | stat. n. | Calisto grannus micrommata |
| Calisto montana | Clench, 1943 | syn. n. | Calisto debarriera |
| Calisto neiba | Schwartz & Gali, 1984 | syn. n. | Calisto debarriera |
| Calisto nubila | Lathy, 1899 | ||
| Calisto obscura | Michener, 1943 | ||
| Calisto phoinix | González, 1987 | stat. n. | Calisto grannus phoinix |
| Calisto pulchella | Lathy, 1899 | ||
| Calisto pulchella darlingtoni | Clench, 1943 | ||
| Calisto raburni | Gali, 1985 | ||
| Calisto sommeri | Schwartz & Gali, 1984 | stat. n. | Calisto grannus sommeri |
| Calisto tasajera | González, Schwartz & Wetherbee, 1991 | ||
| Calisto zangis | (Fabricius, 1775) (Papilio) |
Maximum parsimony (MP) analysis was performed using a heuristic search as implemented in MEGA4 (
Fig. 1 shows the results of the Bayesian Inference analysis (BI). The maximum parsimony analysis revealed a similar topology, but deeper nodes were not strongly supported (bootstrap value < 0.5). Bootstrap values higher than 0.5 are shown on the MP tree (see Supplementary file). For the further analysis and discussion of results we refer to the BI tree. The BI analysis of the tree topology and the Kimura 2-parameter model estimation of genetic distances showed the following results:
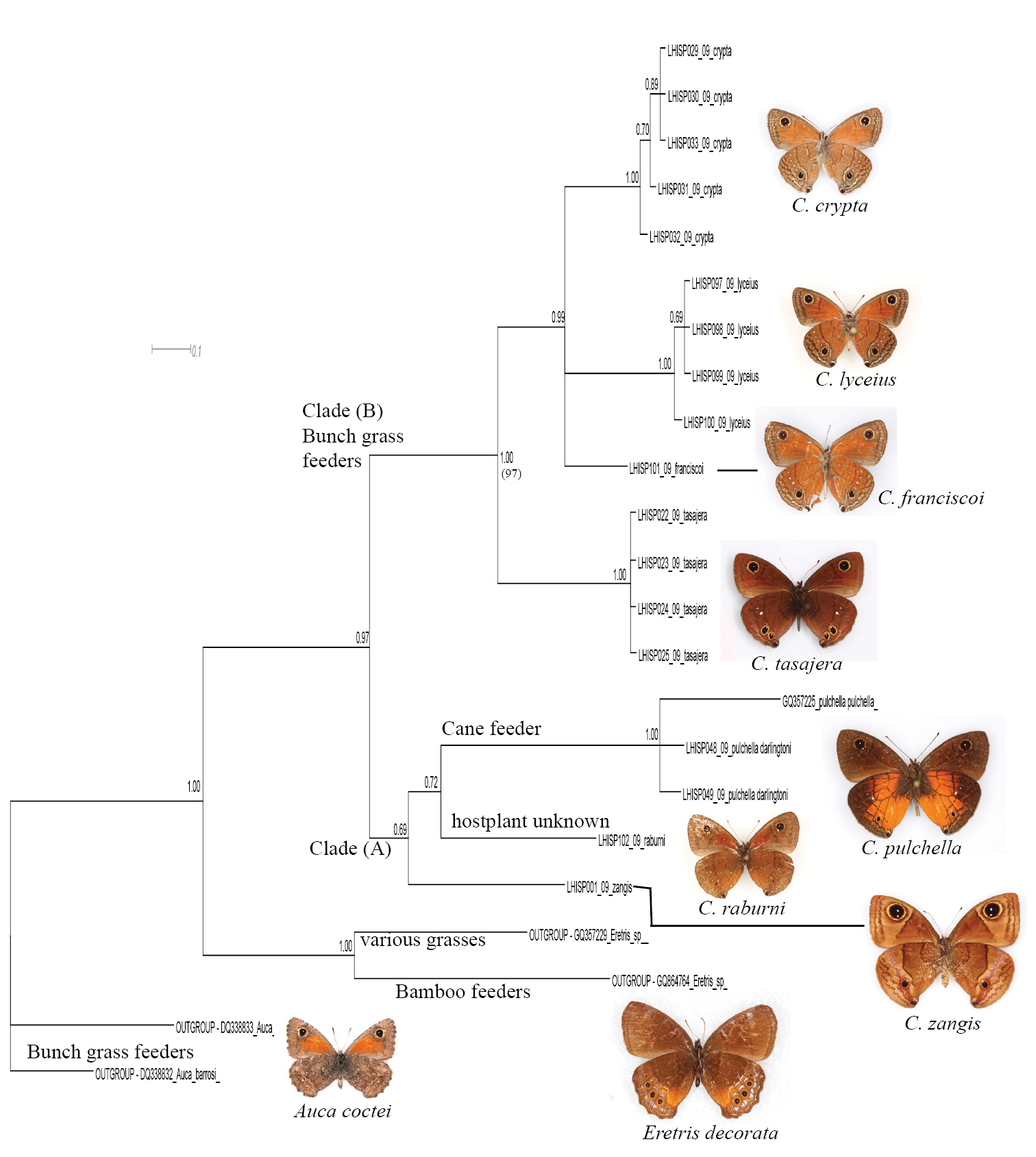
Bayesian inference phylogeny based on 655bp of COI for 111 specimens of the genus Calisto (representing ca. 20 species belonging to 26 named taxa), with outgroups of Eretris and Auca (Nymphalidae: Satyrinae: Prinophilini). The numbers at the nodes indicate posterior probability.
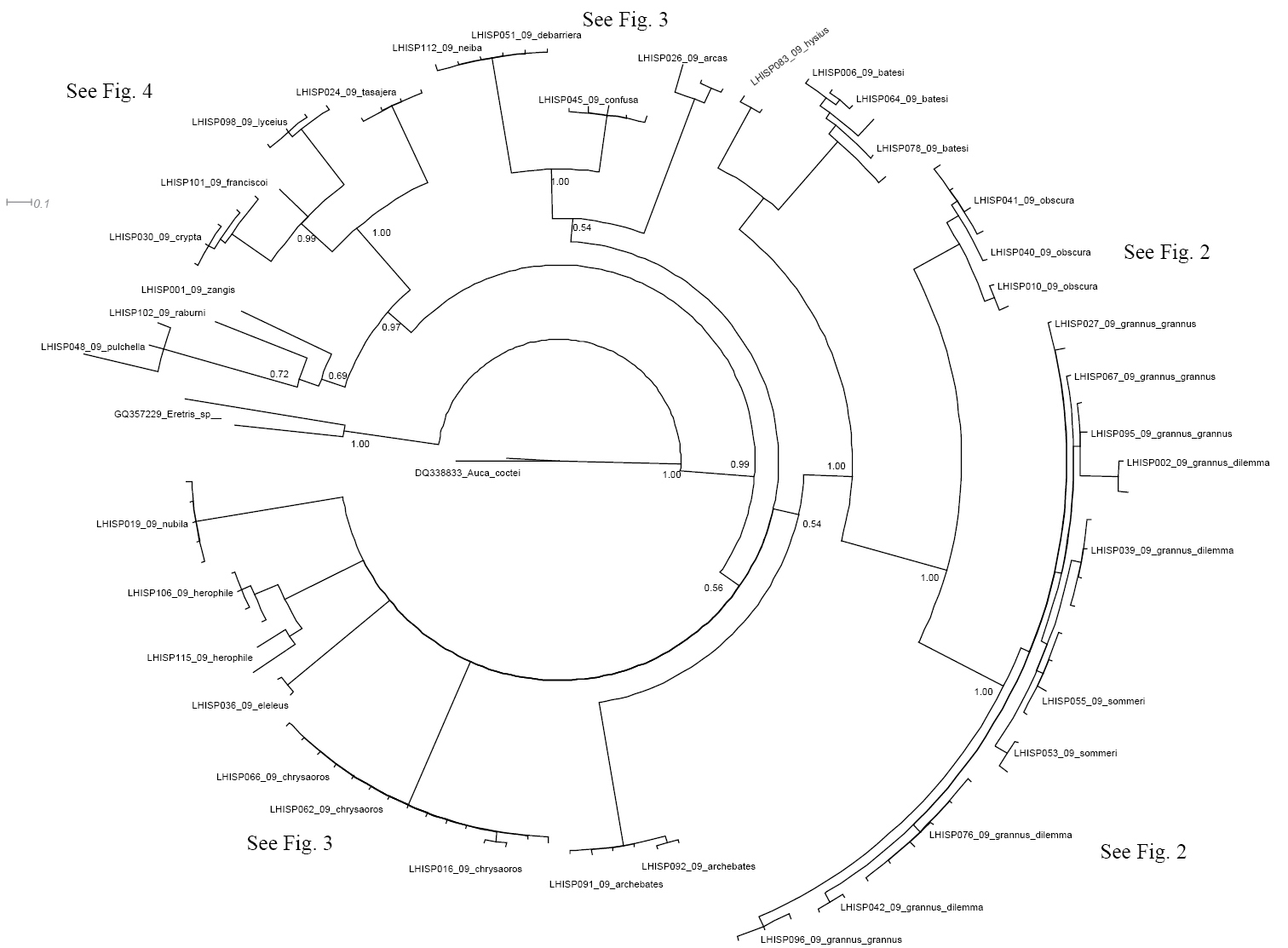
1. The sympatric, superficially similar widespread species Calisto confusa, Calisto obscura and Calisto batesi, which frequently share the same habitat, proved to be extremely divergent. Calisto confusa appear to be related to the morphologically highly derived Calisto arcas Bates, 1939 Fig. 3 (Clades A, B). Calisto obscura, which is found throughout the lowlands and mid-elevations proved to be related to the Calisto grannus species group which is found locally throughout the island at higher elevations (Fig. 2, Clade B). Though the latter clade has Calisto batesi/Calisto hysius species complex as its sister clade (Fig. 2, Clade A), the divergence between Calisto obscura and Calisto batesi is substantial at approximately 9%.
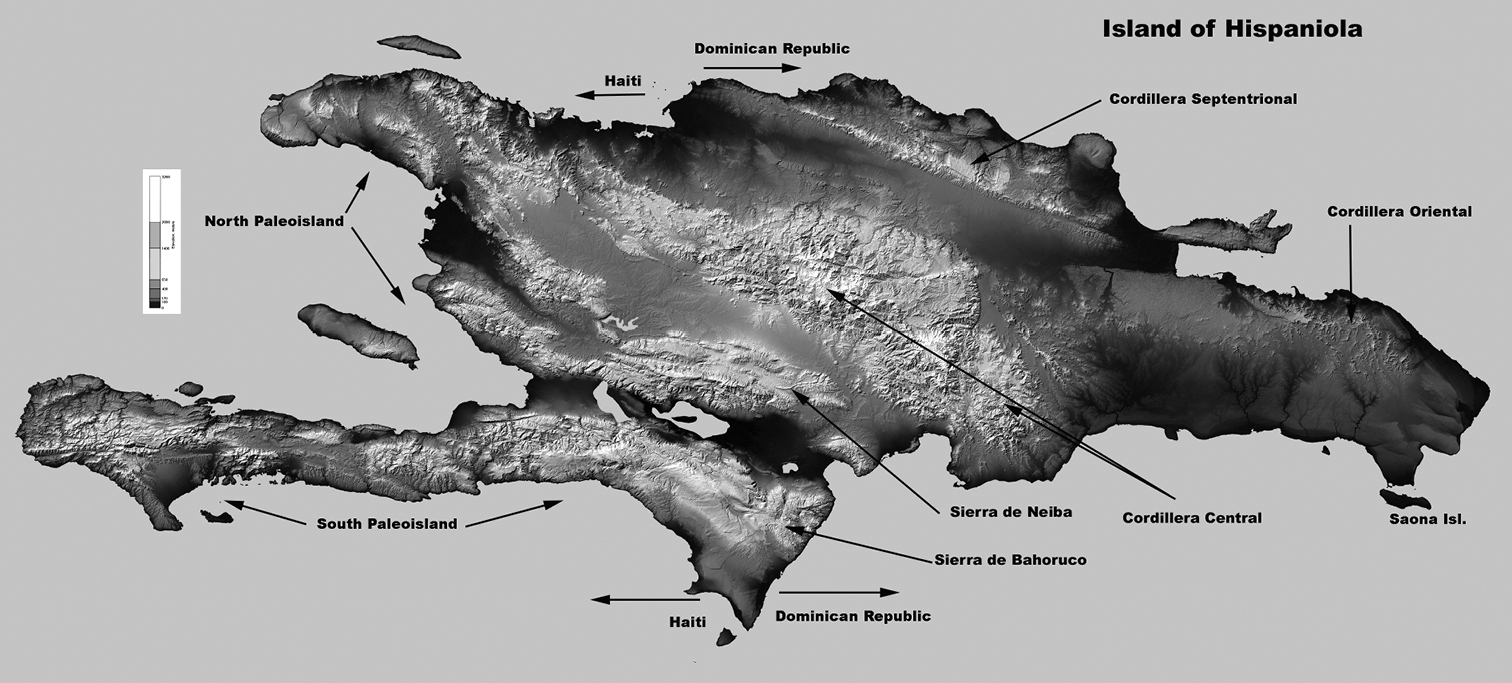
Fragment of the BI tree in Figure 1 with additional information about clades Clade A: Calisto hysius and Calisto batesi are found allopatrically on two Hispaniolan paleoislandsClade B: Calisto obscura is a widespread Hispaniolan species. The Calisto grannus complex is represented by a number of named populations, mostly but not exclusively found in Cordillera Central, the status of which are revised to subspecies in the present study Clade C: Calisto archebates is a local endemic of the southern paleoisland’s Sierra de Bahoruco.
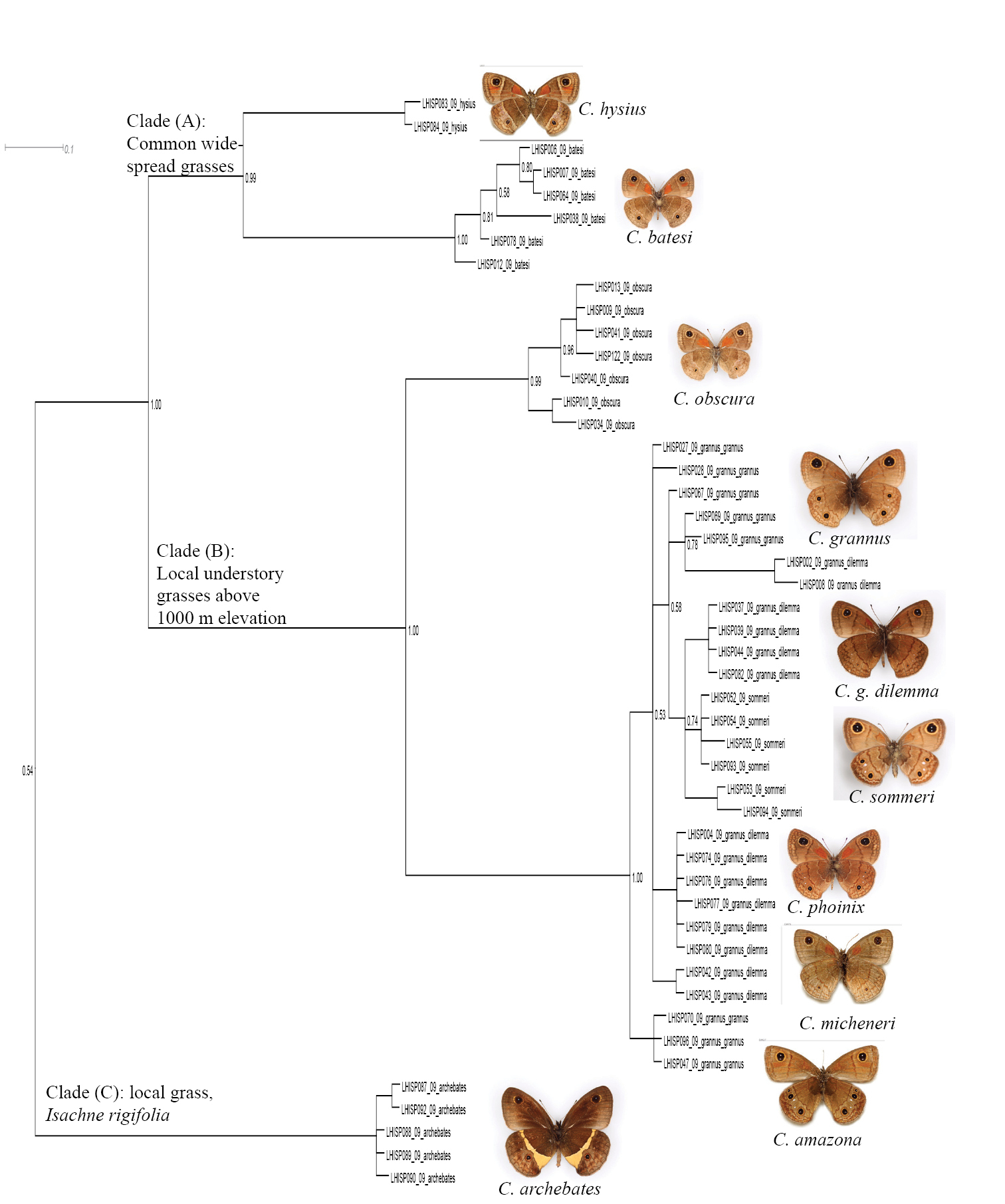
Fragment of the BI tree in Figure 1 with additional information about clades. Clade A: Calisto confusa and Calisto debarriera/Calisto neiba are morphologically similar and sometimes sympatric, though seemingly occupy different elevationsClade B: Calisto arcas is an endemic of Cordillera Central’s Valle Nuevo areaClade C: Calisto chrysaoros is found at high elevations on both southern and northern paleoislands in the refugias associated with climbing bamboo grass Arthrostylidium Clade D: Calisto eleleus is now found extremely locally in the Cordillera CentralClade E: Calisto herophile is distributed on Cuba and Bahamas islands Clade F: Calisto nubila is a Puerto Rican endemic.
2. The allopatric morphologically similar sister species Calisto batesi/Calisto hysius (Fig. 2,
Clade A), whose species status was questionable based on adult
morphology, and whose immature stages are also quite similar (
3. Calisto confusa and Calisto debarriera appeared as two well-separated clusters (Fig. 3, Clade A). Calisto debarriera was originally treated as subspecies of Calisto confusa (
4. Within the Calisto grannus species complex (Fig. 2, clade B), we included at least nine isolated populations from different elevations, which we initially assigned to three taxa: Calisto grannus grannus of high elevations in the Cordillera Central (including a specimen representing the Calisto amazona phenotype), Calisto grannus dilemma (grannus individuals with red discal spot on the underside forewing, which includes such taxa as dilemma, micrommata, dystacta, phoinix, and micheneri) and Calisto sommeri, an isolate from Sierra de Bahoruco. The 28 individuals from these nine populations that are identified on the barcode tree as Calisto grannus grannus, Calisto grannus dilemma and Calisto sommeri show geographic, rather than taxonomic, structure. In other words, individuals cluster within populations, separated from other such clusters by 0.5–1.5%, regardless of the taxonomic name applied. For instance, Calisto sommeri of Sierra de Bahoruco appears as a sister clade to Calisto grannus dilemma from the extreme western portion of Dominican Cordillera Central. The lowland and very common widespread Calisto obscura appears to be a sister taxon to the Calisto grannus species complex, with a divergence of 5–7%.
5. Within the Calisto lyceius species complex (Fig. 4, Clade B), lowland desert isolates such as Calisto crypta, Calisto franciscoi, and Calisto lyceius, despite their superficial morphological similarities, proved to be divergent in their barcodes (ca. 4.5%). Calisto tasajera González, Schwartz & Wetherbee, 1991 proved to be their immediate relative, found at the high elevations.
Fragment of the BI tree in Figure 1 with additional information about clades. The outgroups (Auca - bunch grass feeder from the southern Andes; Eretris - a bamboo-feeding group from Central and South America) and two basal Calisto clades Clade A: Calisto zangis of Jamaica which is aligned with the Hispaniolan Calisto raburni (a rare highly divergent species with an unknown life history) and Calisto pulchella, a well-known sugar cane pest (the native host plant is unknown) Clade B: Calisto tasajera (from the highlands of Cordillera Central) which feeds on Danthonia domingenisis bunch grass and Calisto of the lyceius group feeding on Uniola virgata bunch grass in the Hispaniolan lowlands.
6. A widely-distributed pest of sugar cane, Calisto pulchella (Fig. 4, Clade A) showed a high degree of divergence (3.5%) between its two described subspecies (Calisto pulchella pulchella from the lowlands and Calisto pulchella darlingtoni from the Cordillera Central).
7. Three species endemic to islands other than Hispaniola (Calisto nubila Lathy, 1899, Calisto zangis (Fab., 1775) and Calisto herophile Hübner, 1823) appear to be derived lineages of various Hispaniolan taxa (Fig. 3, Clade D and E; Fig. 4, Clade A). Divergence of these island isolates, though high, does not exceed divergence found within the island of Hispaniola.
8. The maximum divergence within the genus (13.3% between Calisto nubila and Calisto grannus) is almost equivalent to the maximum divergence of Calisto from its distant pronophiline relative Auca from the southern Andes (14.1%), or from its presumed closest relative Eretris (14.4%) (Fig. 4). The average interspecific divergence in Calisto was found to be 10%.
DiscussionAs a result of the present “DNA barcode” analysis, it is
possible to draw a number of taxonomic conclusions (proposed taxonomic
changes are summarized in Table 1). Calisto grannus
represents a recent and incomplete diversification through allopatric
isolation, and for now is best considered as a single species, with Calisto grannus dilemma, Calisto grannus amazona stat. n., Calisto grannus micrommata stat. n., Calisto grannus dystacta stat. n., Calisto grannus phoinix stat. n., Calisto grannus sommeri stat. n., and Calisto grannus micheneri stat. n. representing subspecies. Within the Calisto lyceius complex, lowland desert isolates such as Calisto crypta, Calisto franciscoi, and Calisto lyceius,
despite their superficial morphological similarities, proved to be
sufficiently divergent in their barcodes to confirm their species status
previously postulated based on male genitalia (
Munroe’s view that Calisto confusa and Calisto debarriera
stat. n. are good species is now supported by our DNA data. Munroe
found differences only in aedeagus width/length ratio and immediately
cast doubt on his finding: “No fresh material was examined, and such a
difference might conceivably be the result of distortion of the
preparations.” Munroe examined only four debarriera specimens, but stated that “in support of this evidence it may be noted that the material of debarriera
comes from a limited altitude range, which is entirely contained in
both the altitudinal and geographic range of the widely distributed confusa.”
In other words, Munroe, though only having available a few old
collection specimens, already supposed that he was dealing with two
sympatric taxa. Future workers reduced debarriera to subspecies (e.g.,
The island of Hispaniola, with some key geological features.
Non-Hispaniolan island endemics (Calisto nubila, Calisto zangis and Calisto herophile)
appear to be derived lineages of various Hispaniolan taxa, indicating
several ancient dispersal events from Hispaniola to Puerto Rico, Cuba,
and Jamaica. For instance, Calisto herophile, which occurs in Cuba and the Bahamas, appears to be a product of dispersal from Hispaniola of the widespread polyphagous Calisto confusa or its immediate ancestor. Calisto nubila, endemic to Puerto Rico, which bears morphological similarity to the rare and localized Hispaniolan Calisto eleleus Bates, 1935 (Fig. 3,
Clade D), also most likely have originated by dispersal to Puerto
Rico from the Hispaniolan clade. Divergence of these island isolates,
though great, does not exceed divergence found within the island of
Hispaniola, which suggests that they dispersed from Hispaniola when the
genus was already undergoing diversification. The low diversity of
species on non-Hispaniolan islands as well as the time-frame of Calisto
evolution, suggests that such taxa arrived there by accidental
dispersal, rather than by land bridges or vicariance as hypothesized
previously by
Calisto zangis, along with Calisto pulchella and Calisto raburni
Gali, 1985, are the most morphologically divergent members of the
genus in general wing pattern, male and female genitalic structures,
and in the immature stages (at least for pulchella, for which life history has been studied) (
Our results highlight the usefulness of DNA-barcode
analysis for routine species-level taxonomic work. DNA-barcoding allowed
us to confirm previously observed morphological synapomorphies and test
theories based on morphology and ecology alone. For example, the fact
that the phenotypically divergent species Calisto archebates (Ménétriés, 1832), which has a yellow stripe traversing the hindwing underside, appeared as sister species to the Calisto grannus/Calisto confusa/Calisto batesi complex was already hypothesized based on immature stage morphology (
The evolution of Satyrinae has been linked to the evolution and diversity of grasses (
The butterfly fauna of Hispaniola has evidently been
evolving for many millions of years. For instance, an extinct species
of an extant neotropical genus of Riodinidae is known from Dominican fossil amber, dating from 15–25 Myr (
Examples of habitat diversity on the island of Hispaniola. A Valle de Bao (1920 m elevation) at the foothill of Pico Duarte (3098 m elevation), covered with bunch grass, Danthonia domingenis - a hostplant of Calisto tasajera (top right) B Arid south eastern coastal habitat in Boca de Yuma, Altagracia provides an environment for sea oats, Uniola virgata, and associated Calisto lyceius (top right).
Calisto montana holotype (Museum of Comparative Zoology, Harvard, Massachusetts, USA).
Butterflies, especially grass-feeding butterflies in
such a hurricane-prone area, have thus had many chances to colonize
every possible habitat and island through dispersal. Even though the
genus appears more divergent than most other satyrine genera, it does
not seem to be old enough to be influenced too much by geological events
related to continental movement. Though recognizing the limited ability
of a short DNA strand to give precise time estimates for observed
divergence, most models assume that 1.5–3.5% divergence roughly equates
to one million years of isolation (e. g.,
We thank Keith Willmott for reviewing the manuscript and Alexandra Sourakov for proofreading the early versions. The National Geographic Society funded some of the first author’s fieldwork through Committee for Research and Exploration grant #5717-96. Companions on the field trips, especially Thomas C. Emmel, were instrumental in getting the project to this stage. DNA sequence analysis was carried out at the Canadian Centre for DNA Barcoding through the International Barcode of Life project funded by NSERC, the Ontario Ministry of Research and Innovation and Genome Canada through the Ontario Genomics Institute.