(C) 2011 S. Grozeva. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Eight species belonging to five true bug families were analyzed using DAPI/CMA3-staining and fluorescence in situ hybridization (FISH) with telomeric (TTAGG)n and 18S rDNA probes. Standard chromosomal complements are reported for the first time for Deraeocoris rutilus (Herrich-Schäffer, 1838) (2n=30+2m+XY) and Deraeocoris ruber(Linnaeus, 1758) (2n=30+2m+XY) from the family Miridae. Using FISH, the location of a 18S rDNA cluster was detected in these species and in five more species: Megaloceroea recticornis (Geoffroy, 1785) (2n=30+XY) from the Miridae; Oxycarenus lavaterae (Fabricius, 1787) (2n=14+2m+XY) from the Lygaeidae s.l.; Pyrrhocoris apterus (Linnaeus, 1758) (2n=22+X) from the Pyrrhocoridae; Eurydema oleracea (Linnaeus, 1758) (2n=12+XY) and Graphosoma lineatum (Linnaeus, 1758) (2n=12+XY) from the Pentatomidae. The species were found to differ with respect to location of a 18S rRNA gene cluster which resides on autosomes in Oxycarenus lavaterae and Pyrrhocoris apterus, whereas it locates on sex chromosomes in other five species. The 18S rDNA location provides the first physical landmark of the genomes of the species studied. The insect consensus telomeric pentanucleotide (TTAGG)n was demonstrated to be absent in all the species studied in this respect, Deraeocoris rutilus, Megaloceroea recticornis, Cimex lectularius Linnaeus, 1758 (Cimicidae), Eurydema oleracea, and Graphosoma lineatum, supporting the hypothesis that this motif was lost in early evolution of the Heteroptera and secondarily replaced with another motif (yet unknown) or the alternative telomerase-independent mechanisms of telomere maintenance. Dot-blot hybridization analysis of the genomic DNA from Cimex lectularius, Nabis sp. and Oxycarenus lavaterae with (TTAGG)n and six other telomeric probes likewise provided a negative result.
Karyotypes, meiosis, FISH, 18S rDNA, telomeres, dot-blot, Deraeocoris, Megaloceroea, Nabis, Cimex, Oxycarenus, Pyrrhocoris, Eurydema, Graphosoma, Heteroptera
Fluorescence in situ hybridization(FISH), established in the 1980s, represents a powerful cytogenetic technique for a visualization of specific DNA sequences onto chromosomes, generating detailed chromosome mapping of eukaryote genomes (
To fill this gap and learn more about bug genomes, we applied FISH technique with telomeric (TTAGG)n and 18S rDNAprobes to eight species belonging to 7 genera, 5 families and 2 infrforders: Deraeocoris ruber (Linnaeus, 1758), Deraeocoris rutilus (Herrich-Schäffer, 1838), and Megaloceroea recticornis (Geoffroy, 1785) from the family Miridae, Cimex lectularius Linnaeus, 1758 from the family Cimicidae (all from the infraorder Cimicomorpha); Oxycarenus lavaterae (Fabricius, 1787) from the family Lygaeidae s.l., Pyrrhocoris apterus (Linnaeus, 1758) from the family Pyrrhocoridae, Eurydema oleracea (Linnaeus, 1758) and Graphosoma lineatum (Linnaeus, 1758) from the family Pentatomidae (all from the infraorder Pentatomomorpha). The 18S rDNA location provided the first physical landmark of the genomes of the species studied. The species Deraeocoris ruber and Deraeocoris rutilus were studied here for the first time likewise in terms of their standard chromosomal complements.
In five species, Megaloceroea recticornis, Deraeocoris rutilus, Cimex lectularius, Eurydema oleracea, and Graphosoma lineatum, we used a (TTAGG)n telomeric probe to justify a hypothesis that this telomeric motif is absent in the true bugs (
Additionally, we carried out a dot-blothybridization of the genomic DNA from Cimex lectularius, Nabis sp. and Oxycarenus lavaterae with seven types of telomeric probes, ciliate (TTTTGGGG)n and (TTGGGG)n, nematode (TTAGGC)n, insect (TTAGG)n, shrimp (TAACC)n, vertebrate (TTAGGG)n, and plant (TTTAGGG)n.
Material and methods InsectsAdult males of Deraeocoris ruber, Deraeocoris rutilus, Megaloceroea recticornis, Nabis sp., Cimex lectularius, Oxycarenus lavaterae, Pyrrhocoris apterus, Eurydema oleracea, and Graphosoma lineatum were collected in the vicinities of Plovdiv and Sofia, Bulgaria in 2009-2011 (Table 1). On capture, specimens were immediately fixed in a Carnoy fixative (3 parts of 96% ethanol and 1 part of glacial acetic acid) and stored at 4°C until required.
Material analyzed
| Infraorder, family, and species | Locality in Bulgaria | Date of collection | Number of specimens analyzed |
|---|---|---|---|
| Cimicomorpha | |||
| Miridae | |||
| Deraeocoris ruber | Bulgaria, Western Rhodopes Mts., near Kuklen Vill., 42.032990°N, 024.774537°E, 384 m a.s.l. | 9.06.2009 | 2 |
| Deraeocoris rutilus | Bulgaria, Western Rhodopes Mts., near Kuklen Vill., 42.032990°N, 024.774537°E, 384 m a.s.l. | 8.06.2009 | 2 |
| Megaloceroea recticornis | Bulgaria, Asenovgrad, 42.05977°N, 024.813424°E, 177m a.s.l. | 9.06.2009 | 12 |
| Nabidae | |||
| Nabis sp. | Bulgaria, Sofia, City Center | 15.06.2010 | 4 |
| Cimicidae | |||
| Cimex lectularius | Bulgaria, Sofia, Studentski Grad | 14.10.2010 | 3 |
| Pentatomomorpha | |||
| Lygaeidae | |||
| Oxycarenus lavaterae | Bulgaria, Sofia, City Center, on Tilia sp. | 3.07.2011 | 4 |
| Phyrrocoridae | |||
| Pyrrhocoris apterus | Bulgaria, Sofia, City Center, on Tilia sp. | 3.07.2011 | 2 |
| Pentatomidae | |||
| Eurydema oleracea | Bulgaria, Western Rhodopes Mts., near Progled Vill., 41.68067°N, 024, 70527°E, 1320 m a.s.l. | 9.06.2009 | 2 |
| Graphosoma lineatum | Bulgaria, Thracian Lowland, outflow of Chaya River in Maritsa River, 42.147653°N, 024, 880186°E, 152 m a.s.l. | 8.06.2009 | 3 |
The gonads were dissected out and squashed in a drop of 45% acetic acid. The cover slip was removed using the dry ice. Slides were dehydrated in fresh fixative and air dried. The preparations were first analyzed with a phase contrast microscope at 400x. The best chromosome spreads were used for different staining techniques.
Fluorochrome bandingTo reveal the base composition of C-heterochromatin, staining by GC-specific chromomycin A3 (CMA3) and AT- specific 4-6-diamidino-2-phenylindole (DAPI) was used according to
DNA isolation, PCR amplification, probe generation
Genomic DNA from a male of Pyrrhocoris apterus (Heteroptera, Pyrrhocoridae) was isolated using a Chelex-100 extracted method. FISH using a 18S rRNA gene probe was carried out on the chromosomes of Deraeocoris ruber, Deraeocoris rutilus, Megaloceroea recticornis, Oxycarenus lavaterae, Pyrrhocoris apterus, Eurydema oleracea, and Graphosoma lineatum. FISH using a telomeric (TTAGG)n probe was carried out on the chromosomes of Deraeocoris rutilus, Megaloceroea recticornis, Cimex lectularius, Eurydema oleracea, and Graphosoma lineatum. The target 18S rDNAgene was PCR amplified (primers presented in Table 2) from the genomic DNA of Pyrrhocoris apterus, and labeled by PCR with biotin. Telomere probe (TTAGG)n was PCR amplified and labeled using primers TTAGG_F and TTAGG_R (Table 2) and Rhodamine-5-dUTP (GeneCraft, Germany).
PCR primers used in present study
| Name | Sequence (5‘ – 3‘) |
|---|---|
| 18S_F | ACAAGGGGCACGGACGTAATCAAC |
| 18S_R | CGATACGCGAAT GGCTCAAT |
| Eup_F | TTTTGGGGTTTTGGGGTTTTG |
| Eup_R | CCCCAAAACCCCAAAACCC |
| Prot_F | TTGGGGTTGGGGTTGGGG |
| Prot_R | CCCCAACCCCAACCCCAA |
| Wrm_F | TTAGGCTTAGGCTTAGGCTT |
| Wrm_R | GCCTAAGCCTAAGCCTAAG |
| TTAGG_F | TAACCTAACCTAACCTAACCTAA |
| TTAGG_R | GGTTAGGTTAGGTTAGGTTAGG |
| Shr_F | TAACCTAACCTAACCTAACCTAA |
| Shr_R | GGTTAGGTTAGGTTAGGTTAGG |
| TTAGGG_F | CCCTAACCCTAACCCTAACCCTAACCCTAA |
| TTAGGG_R | TTAGGGTTAGGGTTAGGGTTAGGGTTAGGG |
| Plnt_F | TTTAGGGTTTAGGGTTTAGGG |
| Plnt_R | CCCTAAACCCTAAACCCTAAA |
In situ hybridization was performed as described by
Genomic DNA from Cimex lectularius, Oxycarenus lavaterae and Nabis sp. was isolated using NucleoSpin Tissue Kit (Macherey-Nagel, Germany) or a standard Phenol/Chloroform nucleic acid extraction protocol. Telomere probes of ciliate (TTTTGGGG)n and (TTGGGG)n, nematode (TTAGGC)n, insect (TTAGG)n, shrimp (TAACC)n, vertebrate (TTAGGG)n and plant (TTTAGGG)n were PCR amplified using primers labeled with biotin and presented in Table 2.
About 20 ηg of isolated DNA after denaturation was added drop wise to Hybond N+ nylon membranes (Amersham, Biosciences). Hybridizations were carried out over night in hybridization mixture containing about 100-200 ηg of labeled probe, 50% formamide, 4×SSC, 0.5% (w/v) SDS and 10 µg salmon-sperm DNA at 40 °C. Membranes were washed two times in 2x SSC/0.1% SDS (10 min each) at RT and two times in 0.2x SSC/0.1% SDS (10 min each) at RT (10 min each). Detection procedure was performed according to the Biotin Chromogenic Detection Kit protocol (Fermentas).
Microscopy and imagingChromosome preparations were analyzed under a Leica DM 4000B microscope with a 100x objective. Fluorescence images were taken with a Leica DFC 350 FX camera using Leica Application Suite 2.8.1 software with an Image Overlay module. The preparations were stored partly at Institute of Biodiversity and Ecosystem Research, BAS in Sofia and partly at the Zoological Institute, RAS in St Petersburg.
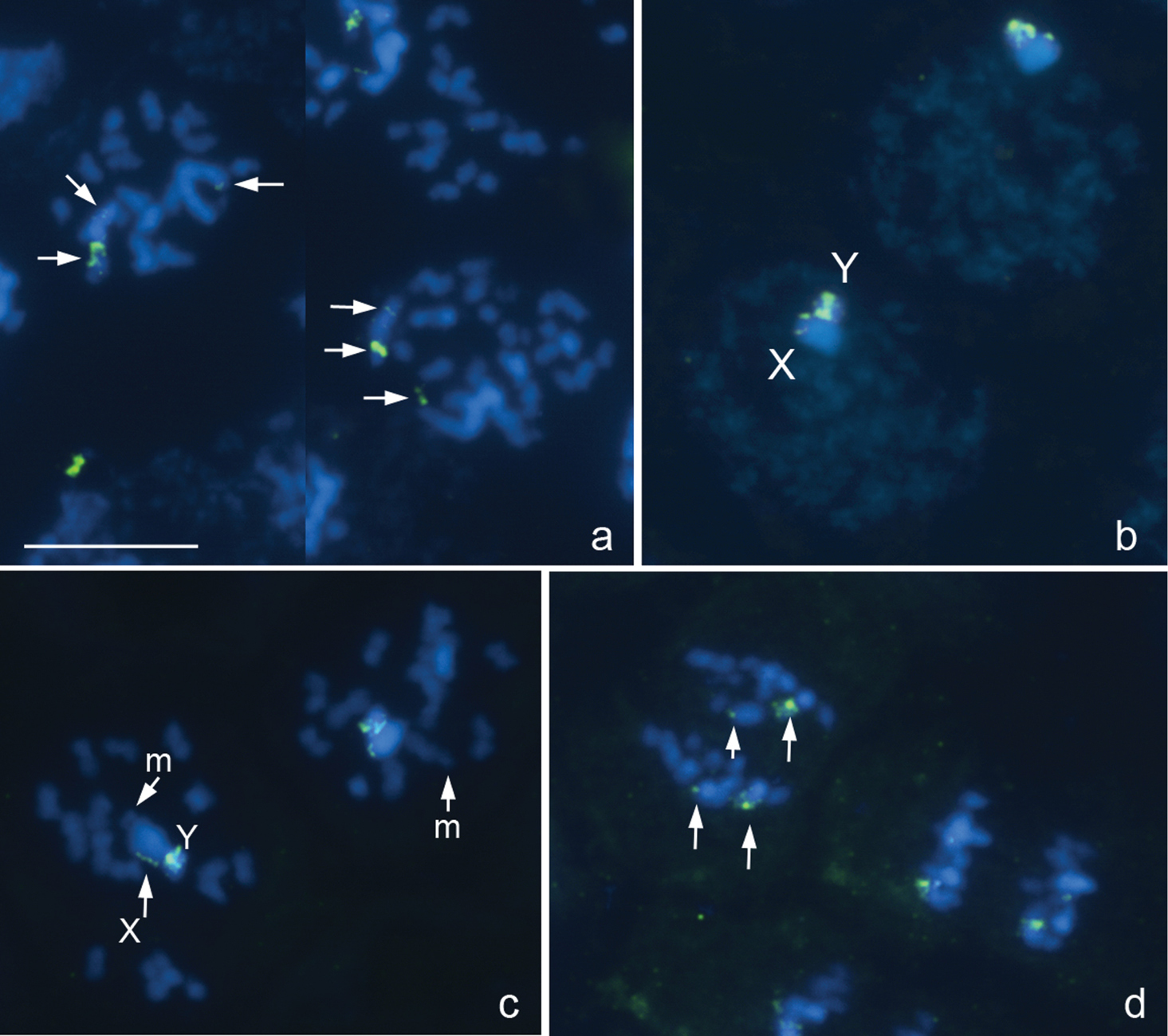
Results MiridaeDeraeocoris ruber Linnaeus, 1758, 2n=30+2m+XY
Fig. 1a–d
The karyotype is described here for the first time. There are 16 bivalents, including a pair of very small and negatively heteropycnotic chromosomes taken as a pair of m-chromosomes, and the univalent X and Y chromosomes which are largest and smallest chromosomes in the set, respectively (Fig. 1a–d). Diplotene and diakinesis stages were not detected, and bivalents displayed no chiasmata since meiosis is achiasmate. 18S rRNA genes were mapped on both sex chromosomes, the signals being more intensive on the Y. At spermatogonial metaphases, a number of very small intercalary signals could be in addition seen on the X (Fig. 1a).
Deraeocoris ruber, 2n=30+2m+XY. a spermatogonial prometaphase b early prophase c prometaphase I d anaphase II. FISH with an 18S rDNA probe. Arrowed are 18S rDNA clusters a, d Bar equals 10 μm.
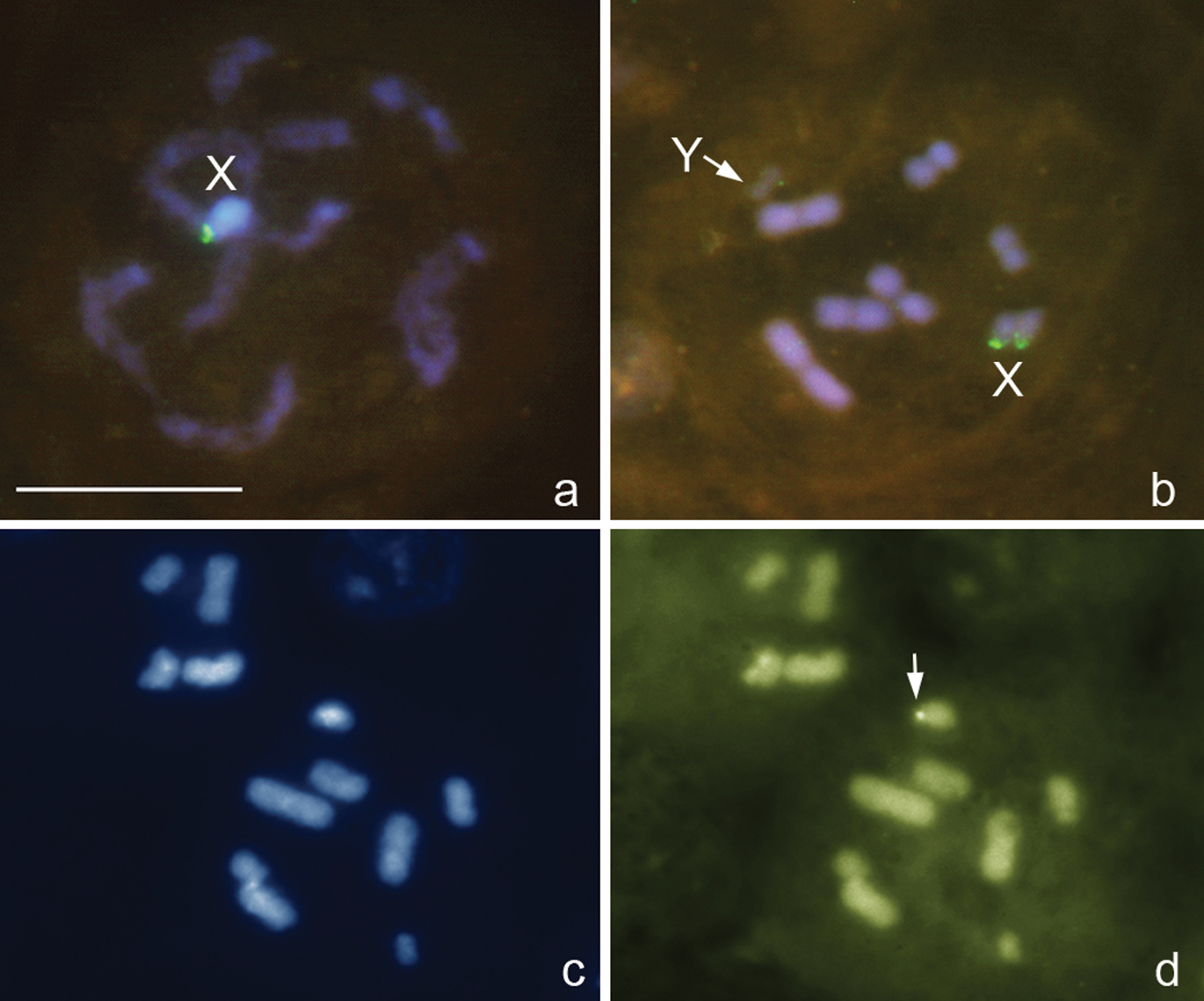
Deraeocoris rutilus (Herrich-Schäffer, 1838), 2n=30+2m+XY
Fig. 2a, b
The karyotype is described here for the first time. It is much like that described above for Deraeocoris ruber. Likewise, at PMI, there are 16 autosomal bivalents and the univalent X and Y chromosomes. One of the bivalents is very small, negatively heteropycnotic pair of m-chromosomes (Fig. 2a, b). The X is the largest and the Y is one of the smallest chromosomes in the set (excluding the m-chromosomes); autosomal bilvalents constitute a decreasing size raw. Diplotene and diakinesis stages were not observed, and meiosis was considered achiasmate of a collochore type. FISH with an 18S rDNAprobe produced two local signals placed near a telomeric region of the X chromosome and in addition a huge cluster of signals attached to the X; Y chromosome carried no signal (Fig. 2a, b). FISH with a TTAGG probe produced prominent hybridization signals (Fig. 2a, b, arrowed), which were occasionally located on the same chromosomes however most likely did not indicate the telomeres.
Deraeocoris rutilus, 2n=30+2m+XY. a, b prometaphase I. FISH with 18S rDNA and (TTAGG)n probes. Arrowed are signals after using a TTAGG-probe. Bar equals 10 μm.
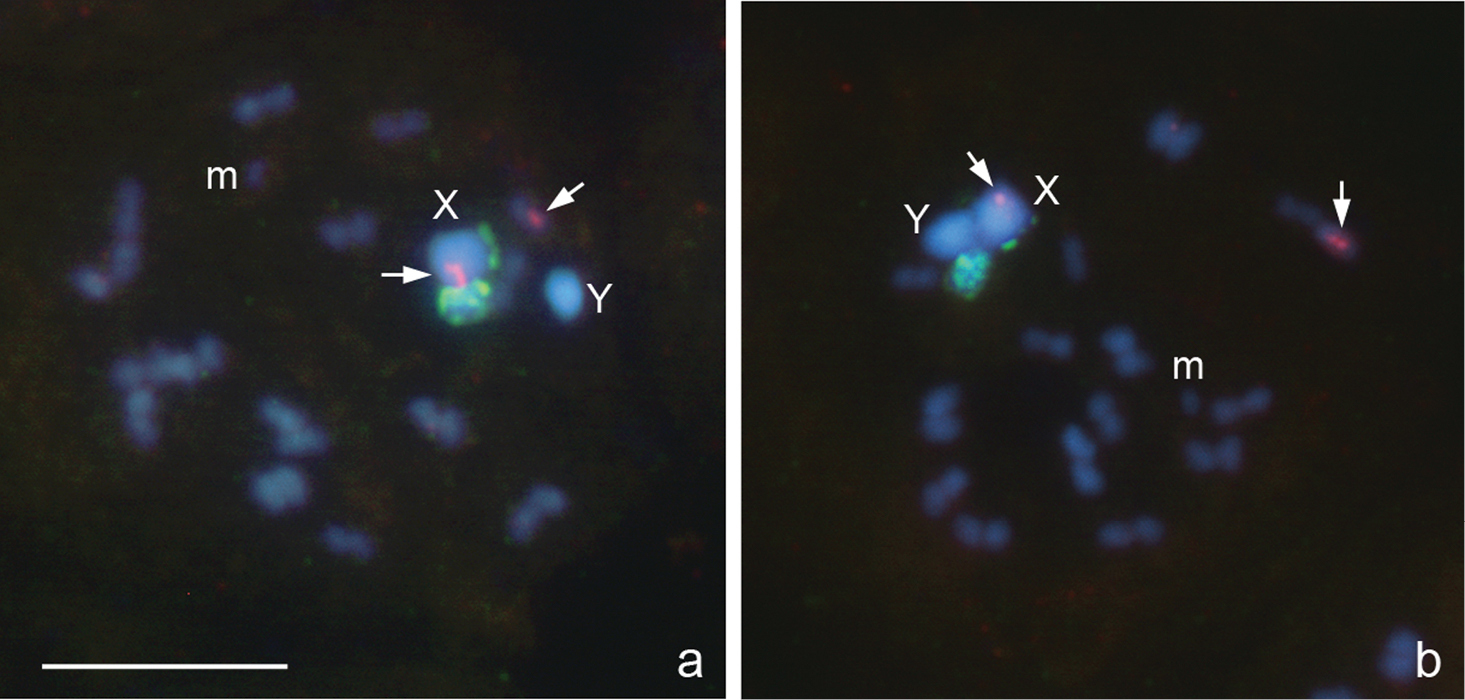
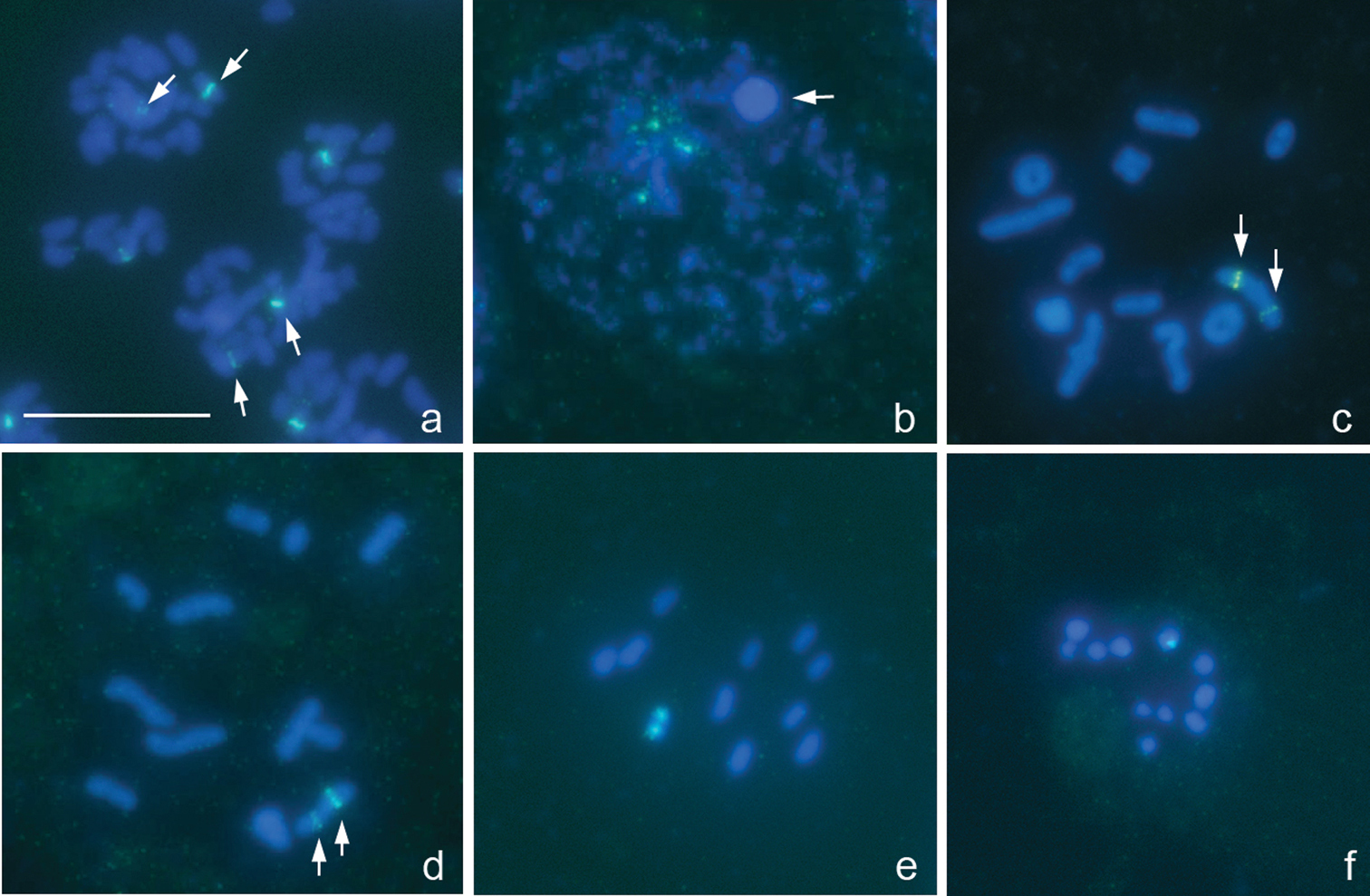
Megaloceroea recticornis (Geoffroy, 1785), 2n=30+XY
Fig. 3a–f
From the counts of 23 plates at prometaphase I (PMI), 15 autosomal bivalents and two univalent sex chromosomes, X and Y, were detected suggesting that this species displays 2n=30+XY (Fig. 3a-c) in contrast to 2n=32+XY previously reported for this species in England (
Megaloceroea recticornis, 2n=30+XY. a, b, d condensation stage; c, e prometaphase I; f metaphase II. FISH with 18S rDNA a, b, d–f and (TTAGG)nf probes. Bar equals 10 μm.
Cimex lectularius Linnaeus, 1758, 2n=26+X1X2Y
not figured
This study confirms that Cimex lectularius display 2n=26+X1X2Y as it was repeatedly reported previously (see
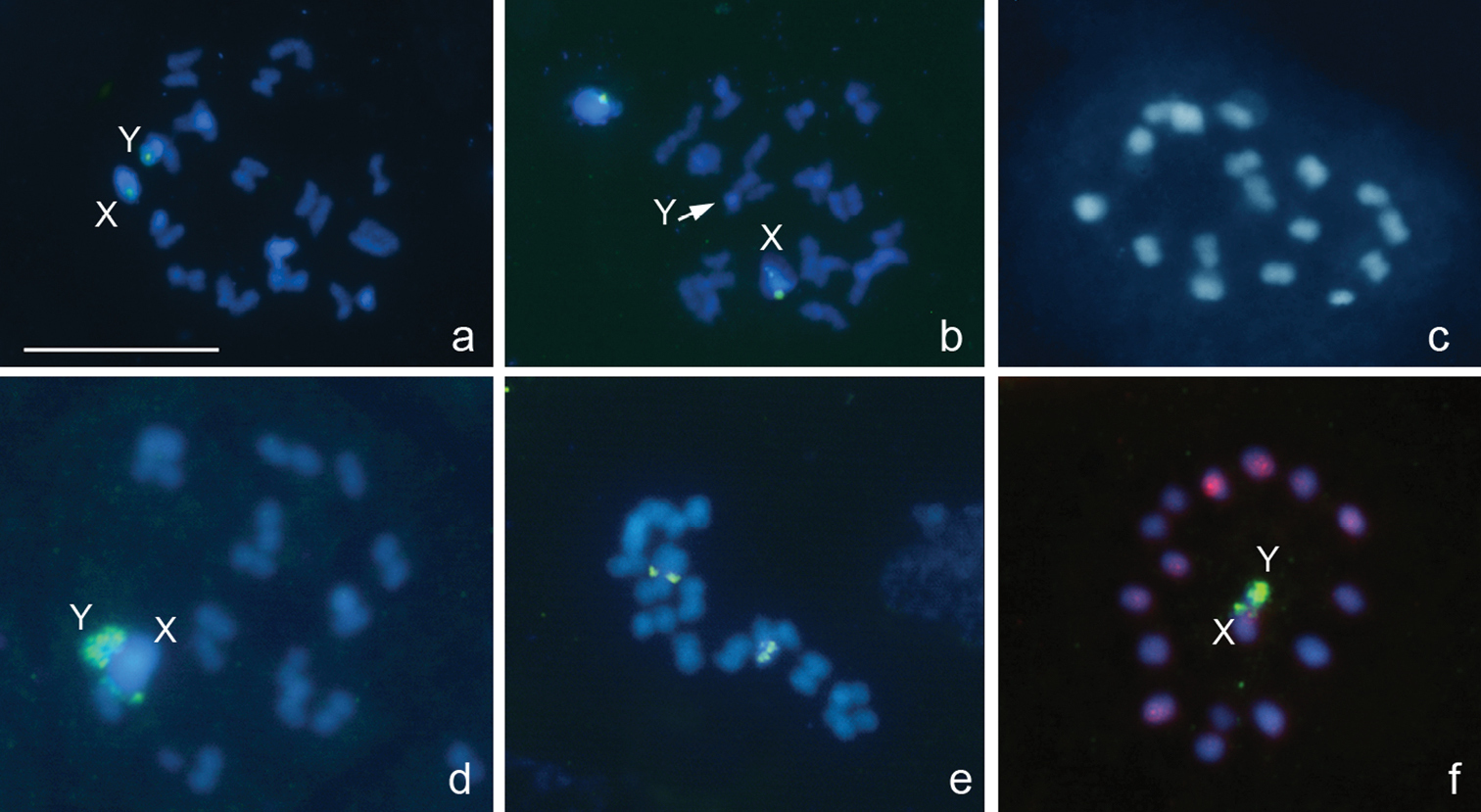
Oxycarenus lavaterae (Fabricius, 1787), 2n=14+2m+XY
Fig. 4a, b
In accordance with earlier published data (
Oxycarenus lavaterae, 2n=14+2m+XY. a spermatogonial metaphase b metaphase I. FISH with a 18S rDNA probe. Bar equals 10 μm.
Pyrrhocoris apterus (Linnaeus, 1758), 2n=22+X
Fig. 5a–f
In accordance with previously published data (
Pyrrhocoris apterus, 2n=22+X. a spermatogonial prometaphase b early prophase (arrowed is the sex chromosome body) c, d prometaphase I e metaphase II f telophase II. FISH with an 18S rDNA probe. Arrowed are 18S rDNA clusters. Bar equals 10 μm.
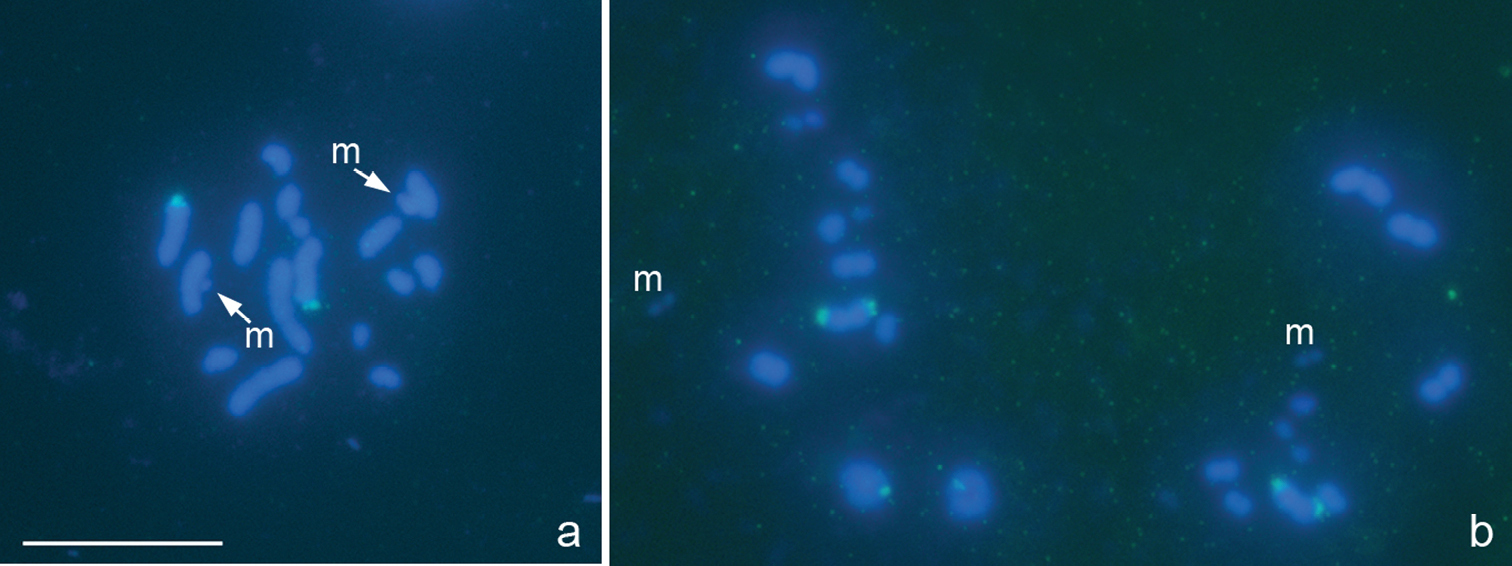
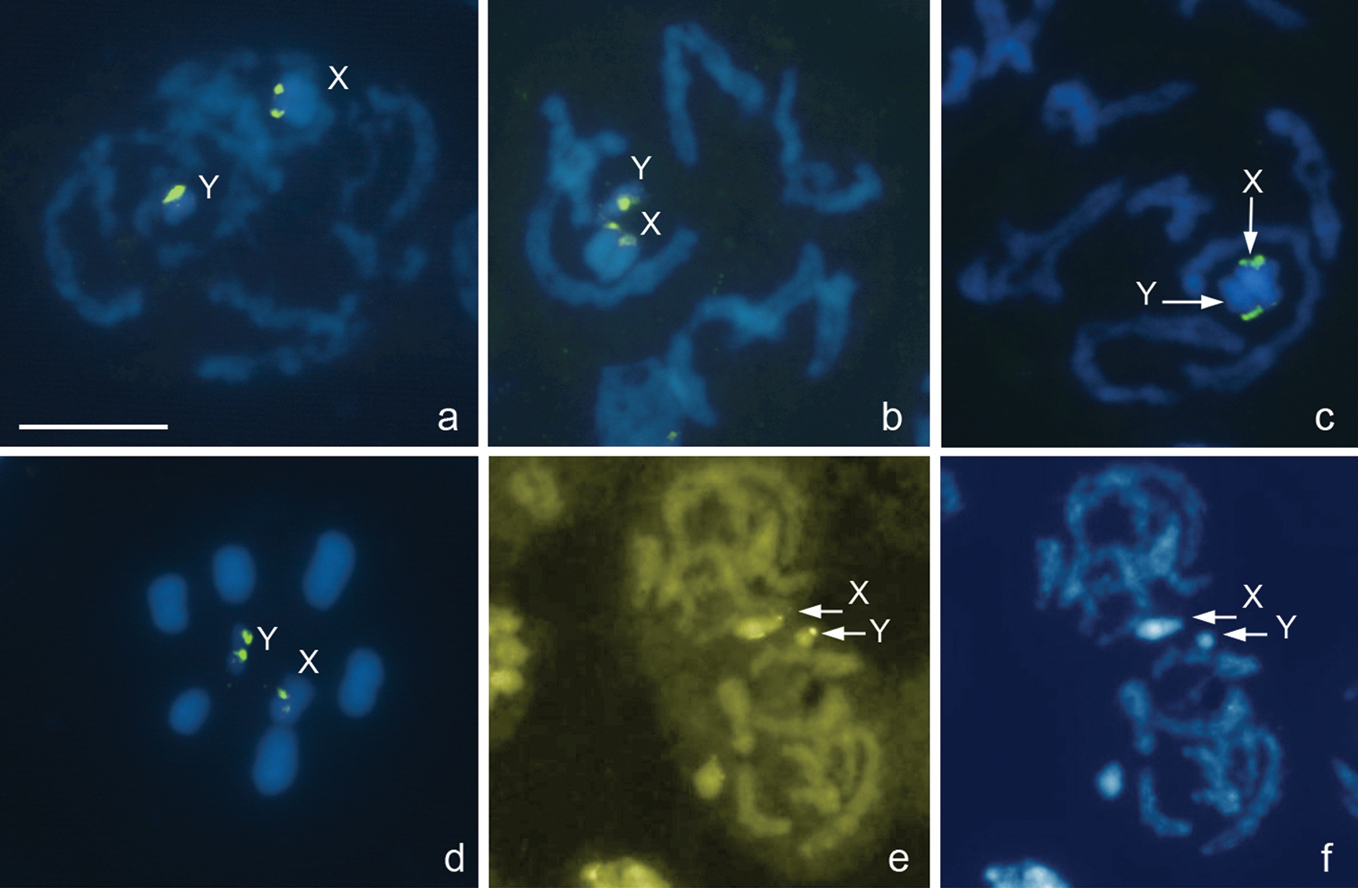
Eurydema oleracea (Linnaeus, 1758), 2n=12+XY
Fig. 6a–f
This study confirms that Eurydema oleracea has 2n=12+XY as previously reported by other researchers (
Eurydema oleracea, 2n=12+XY. a-c, e, f different prophase stages d metaphase I. FISH with a 18S rDNA probe a-d and CMA3 /DAPI e/f-staining. Bar equals 10 μm.
Graphosoma lineatum (Linnaeus, 1758), 2n=12+XY
Fig. 7a–d
The karyotype of 2n=12+XY discovered here in Graphosoma lineatum is in accordance with that published previously for other populations of this species (
Graphosoma lineatum, 2n=12+XY. a prophase stage b–d metaphase I. FISH with 18S rDNAand (TTAGG)n probes a, b and DAPI c and CMA3 d-staining. Arrowed d is a CMA3-positive signal on the X. Bar equals 10 μm.
Dot-blot hybridization of the genomic DNA from Cimex lectularius, Nabis sp. and Oxycarenus lavaterae was performed using seven telomeric probes, ciliate (TTTTGGGG)n and (TTGGGG)n, nematode (TTAGGC)n, insect (TTAGG)n, shrimp (TAACC)n, vertebrate (TTAGGG)n and plant (TTTAGGG)n. All the experiments provided no hybridizing bands clearly suggesting some other molecular composition of telomeres in true bugs.
Discussion Standard chromosomal complementsWe studied standard chromosomal complements of eight species from 6 genera and 5 families of the true bug infraorders Cimicomorpha (Deraeocoris rubber, Deraeocoris rutilus, Megaloceroea recticornis, Cimex lectularius) and Pentatomomorpha (Oxycarenus lavaterae, Pyrrhocoris apterus, Eurydema oleracea, Graphosoma lineatum). Our study confirms the previously published information (see Results and Table 3 for the references) that Cimex lectularius (Cimicidae) displays 2n=26+X1X2Y; Pyrrhocoris apterus (Pyrrhocoridae) – 2n=22+X; Oxycarenus lavaterae (Ligaeidae) – 2n=14+2m+XY; Eurydema oleraceaand Graphosoma lineatum (Pentatomidae) – 2n=12+XY. On the other hand,
Chromosomal complements and 18S DNA locations in the species studied
| Infraorder, family, and species | 2n (♂) | Karyotype formula | 18S rDNA location | Published data on karyotype |
|---|---|---|---|---|
| Cimicomorpha | ||||
| Miridae | ||||
| Deraeocoris ruber | 34 | 2n=30+2m+XY | X and Y chromosomes | Absent |
| Deraeocoris rutilus | 34 | 2n=30+2m+XY | X chromosome | Absent |
| Megaloceroea recticornis | 32 | 2n=30+XY | X and Y chromosomes | 2n=32+XY ( |
| Cimicidae | ||||
| Cimex lectularius | 29 | 2n=26+X1X2Y | X1 and Y chromosomes* | 2n=26+X1X2Y ( |
| Pentatomomorpha | ||||
| Ligaeidae | ||||
| Oxycarenus lavaterae | 18 | 2n=14+2m+XY |
A pair of larger autosomes | 2n=14+2m+XY ( |
| Phyrrocoridae | ||||
| Pyrrhocoris apterus | 23 | 2n=22+X | A pair of larger autosomes | 2n=22+XX/X0 ( |
| Pentatomidae | ||||
| Eurydema oleracea | 14 | 2n = 12+XY | X and Y chromosomes | 2n=12+XY ( |
| Graphosoma lineatum | 14 | 2n = 12+XY | X chromosome | 2n=12+XY ( |
Male meiosis
In all the seven species studied in this work, the first meiotic division is reductional for the autosomes and equational for the sex chromosomes, and vice versa – the second division is equational for the autosomes and reductional for the sex chromosomes. Such a behavior of sex chromosomes in male meiosis, or “post-reduction”, as it is called, represents one of the unique cytogenetic characters of the Heteroptera being inherent in most bug species (
One of the distinctive properties of the true bug meiosis is a specific spatial arrangement of metaphase plates known as radial ones. Either at both metaphases, MI and MII, or at only one of those, the autosomes (either as bivalents at MI or as univalents at MII) form a ring with the sex chromosomes (either as univalents at MI or as a pseudo-bivalent at MII) lying in its center (
The nucleolus represents a subnuclear compartment of eukaryotic cells in which the synthesis of ribosomal RNA (rRNA) and formation of ribosomes take place (
In Heteroptera, physical location of genes remains very poorly sampled (and the data available concern only ribosomal genes), mainly with sporadic sampling of a few select species. Out of more than 40, 000 described species (
In this study we have characterized the chromosomal locations of 18S RNA genes in seven species from six genera of the families Miridae (Deraeocoris ruber, Deraeocoris rutilus, Megaloceroea recticornis), Lygaeidae (Oxycarenus lavaterae), Pyrrhocoridae (Pyrrhocoris apterus), and Pentatomidae (Eurydema oleracea, Graphosoma lineatum). Data on all the species (as well as those on the whole families Miridae and Lygaeidae) were obtained for the first time bringing thus the total number of the species and genera studied to 30 and 18, respectively. The species were shown to exhibit different patterns of rDNA location. Some of the species showed their ribosomic cistrons located on sex chromosome, either on the X (Deraeocoris rutilus) or on both X and Y(Deraeocoris ruber, Megaloceroea recticornis, Eurydema oleracea, Graphosoma lineatum) whereas in other species (Oxycarenus lavaterae, Pyrrhocoris apterus) they were located on a pair of autosomes.
The location of 18S rDNAappeared different within the taxa, in which rDNA sequences were mapped in more than one species as in the mirid genus Deraeocoris, where Deraeocoris rutilus displayed rDNA clusters concentrated on the X, but Deraeocoris ruber on both X and Y chromosomes. These findings give no way of any inferences especially as a wide variation of chromosomal location for the major rDNA has been observed in different bug taxa, including variations between the co-generic species. For example, the genus Triatoma Laporte, 1832 from the subfamily Triatominae (Reduviidae), which is one of the most studied bug groups, clearly shows the interspecific variation (
The pentanucleotide sequence (TTAGG)n is known as the commonest and most likely an ancestral DNA motif of insect telomeres (
The absence of (TTAGG)n telomeric repeat in the phylogenetically distant groups within the Heteroptera strengthens thus the view (
Dot-blot hybridization of the genomic DNA from bug species with seven types of telomeric probes, ciliate (TTTTGGGG)n and (TTGGGG)n, nematode (TTAGGC)n, insect (TTAGG)n, shrimp (TAACC)n, vertebrate (TTAGGG)n and plant (TTTAGGG)n, yielded negative results and did not provide hence any answer of the question which is the telomere repeat sequence in bug chromosomes.
This study was supported financially by the Russian Foundation for Basic Research (grant 11-04-00734) and programs of the Presidium of the Russian Academy of Sciences “Gene Pools and Genetic Diversity” and “Origin of the Biosphere and Evolution of Geo-biological Systems” (for VK and BA) and by National Scientific Fund of Bulgarian Ministry of Education, Youth and Science (DO-02-259/08) (for SG). We express our thanks to Nikolay Simov (NMNH, Sofia) who provided us the insects for present study.