(C) 2011 J.A. Bitencourt. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Cytogenetic analyses using C-banding and chromosomal digestion by several restriction enzymes were carried out in four populations (named A, B, C and D) of Hypostomus prope unae (Loricariidae, Hypostominae) from Contas river basin, northeastern Brazil. These populations share 2n=76 and single NORs on the second metacentric pair but exclusive karyotype forms for each locality. Populations A and B presented conspicuous terminal and interstitial heterochromatic blocks on most of acrocentric chromosomes and equivalent to NORs with differences in both position and bearing pair. Population D showed evident marks at interstitial regions and interspersed with nucleolar region while population C presented interstitial and terminal heterochromatin segments, non-coincident with NORs. The banding pattern after digestion with the endonucleases Alu I, Bam HI, Hae III and Dde I revealed a remarkable heterogeneity within heterochromatin, allowing the identification of distinctive clusters of repeated DNA in the studied populations, besides specific patterns along euchromatic regions. The analysis using restriction enzymes has proved to be highly informative, characterizing population differences and peculiarities in the genome organization of Hypostomus prope unae.
C-banding, heterochromatin, ichthyofauna, restriction enzymes
Restriction enzymes (RE) represent a powerful tool for studies about DNA organization (
In spite of the intensive application of restriction enzymes in chromosomal analyses of several animal groups (
Within the genus Hypostomus Lacépède, 1803, heterochromatin can be associated to heteromorphic chromosomes (
The goal of the present work was to analyze comparatively metaphase chromosomes of Hypostomus prope unae (Steindachner, 1878) by C-banding and RE digestion in order to refine previous cytogenetic studies (
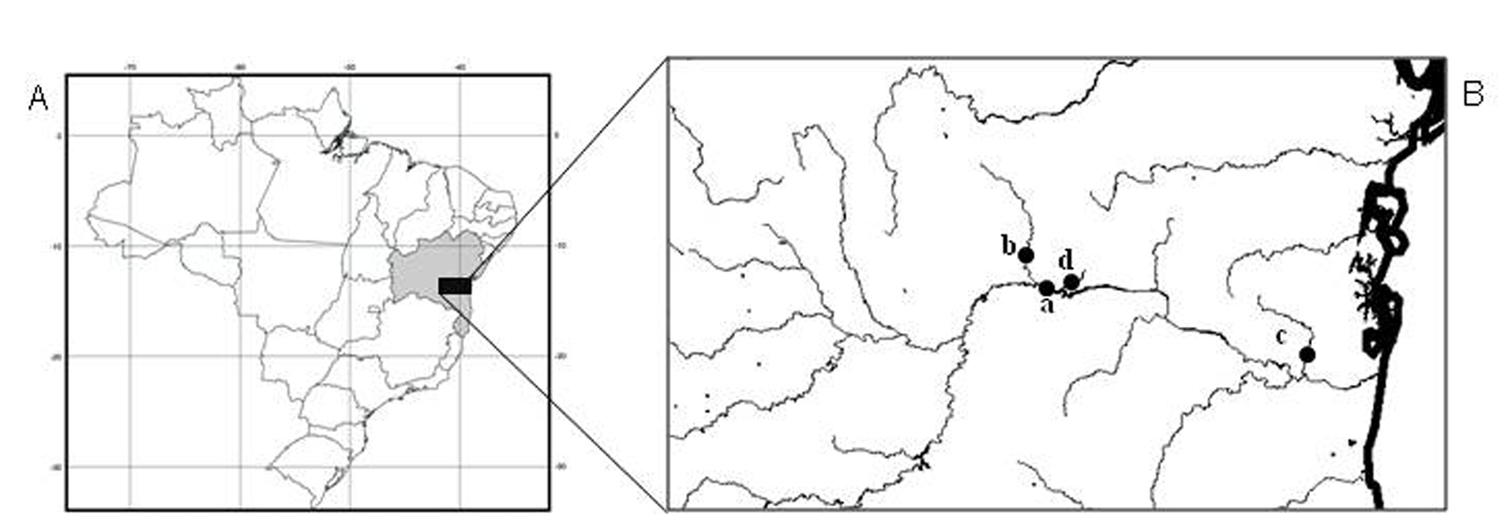
Forty-six specimens of Hypostomus prope unae from four collection sites in Contas river basin were analyzed, being 10 (3 males, 2 females and 5 immature) from the main channel of Contas river (13°51'51"S, 40°04'54"W), 10 (6 males, 1 female and 3 immature) from Preto do Costa river (13°45'84"S, 39°56'47"W), 15 (9 males and 6 immature) from Oricó river (14°08'03"S, 39°21'30"W), and 11 (4 males, 4 females and 3 immature) from Preto do Criciúma river (13°55'45"S, 39°57'57"W) (Fig. 1).
A–B Collection sites A Map of Brazil, highlighting the state of Bahia in northeastern region; B Contas river basin and respective sampling sites: a- Contas river, b- Preto do Costa river, c- Oricó river, d- Preto do Criciúma river.
Voucher specimens were identified by Dr. Claudio Zawadski from Universidade Estadual de Maringá (UEM) and deposited in the fish collection at NUPELIA – UEM, Maringá, PR, Brazil (NUP 9811, 9814). These four populations are referred as A, B, C and D, respectively.
Metaphase chromosomes were obtained from kidney cells as described by
C-positive heterochromatin was detected according to
List of restriction endonucleases (RE) used on the chromosomal preparations of Hypostomus prope unae, with their respective restriction sites and optimum concentrations and incubation periods obtained in the present work.
| Endonucleases | Restriction site | Concentration | Incubation |
|---|---|---|---|
| Alu I | (5’- AG ↓ CT - 3’) | 0.4 U/ µl | 4h |
| Bam HI | (5’- G ↓ GATCC - 3’) | 0.5 U/ µl | 15h |
| Hae III | (5’-GG ↓ CC - 3’) | 0.6 U/ µl | 14h |
| Dde I | (5’- C ↓ TNAG – 3’) | 2 U/ µl | 4h |
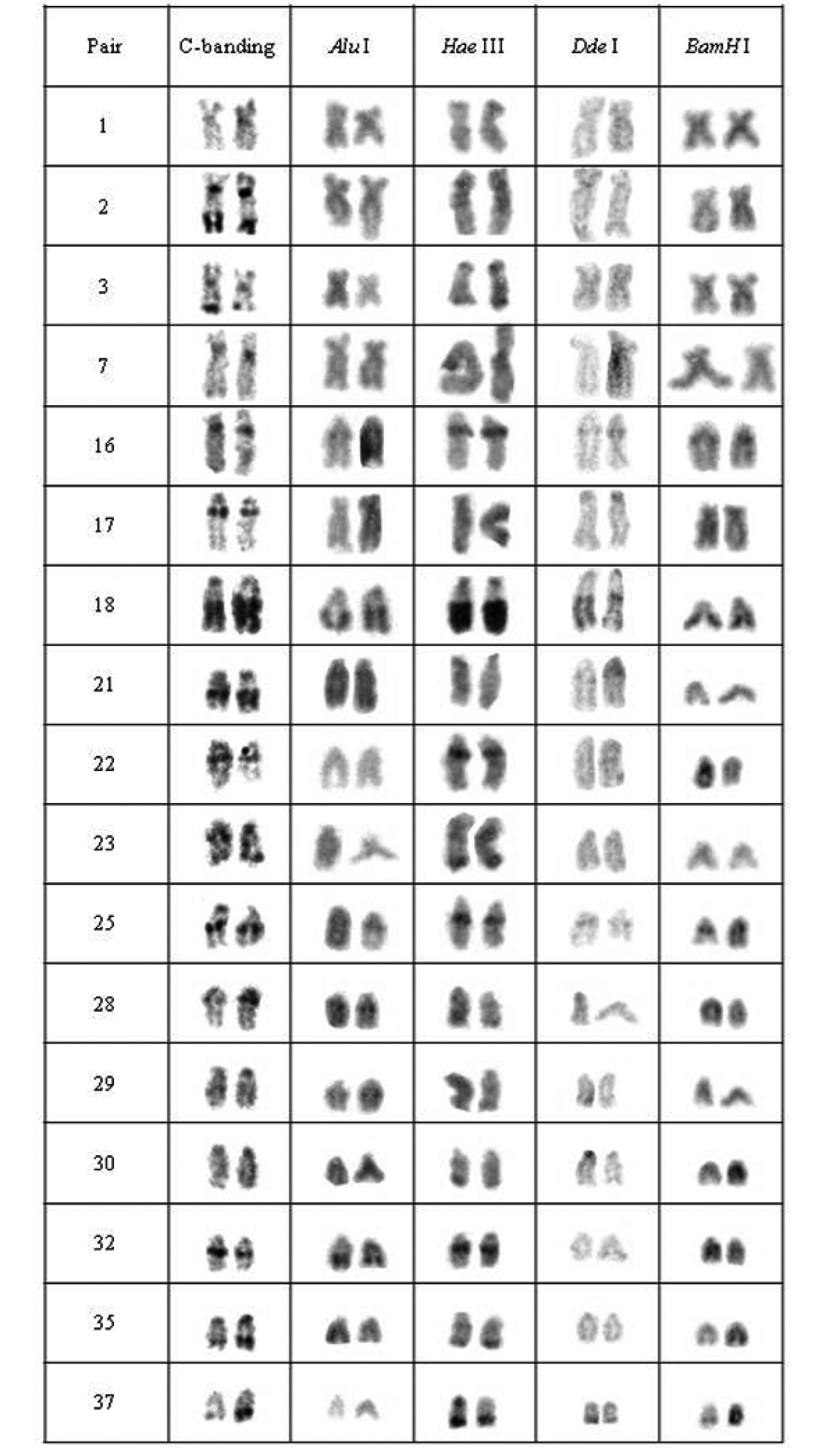
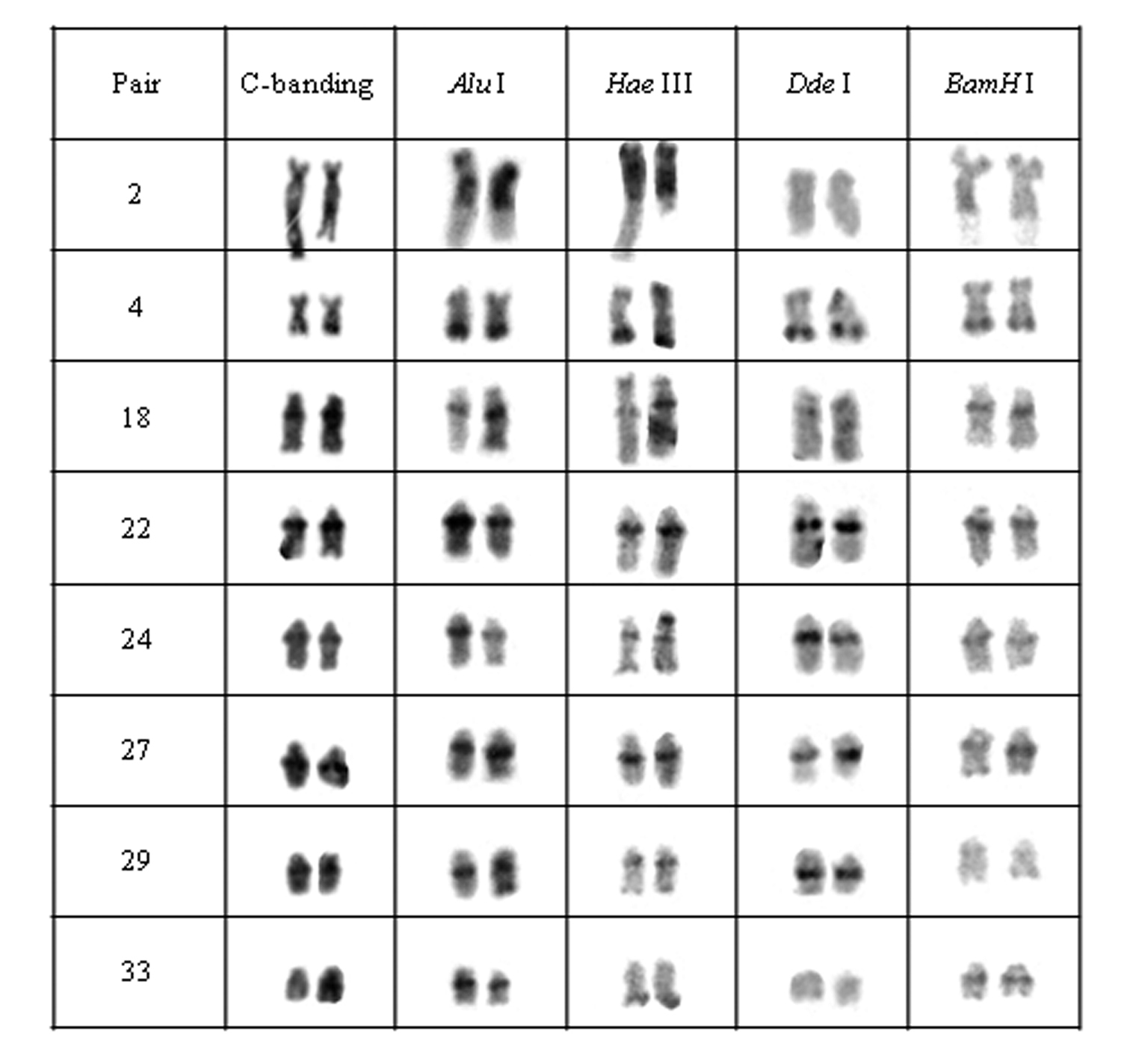
The specimens from all analyzed populations presented a modal chromosomal number of 2n=76 and distinct karyotype formulae, as follows: 12m+16sm+48st/a (FN= 104) for specimens from population A, 12m+20sm+44st/a (FN=108) for specimens from population B, 10m+14sm+52st/a (FN=100) for individuals from population C and 10m+20sm+46st/a (FN= 106) for those from population D. Furthermore, distinctive patterns of heterochromatin distribution were detected by C-banding. Although populations A and B bear conspicuous terminal and interstitial marks in 17 chromosomal pairs as well as centromeric and NOR-associated heterochromatin, they differ in relation to C-bands position or bearing pair (Figs 2, 3).
Chromosomal pairs from population A of Hypostomus prope unae showing the C-positive heterochromatin and banding pattern after digestion with the restriction endonucleases: Alu I, Hae III, Dde I and Bam HI.
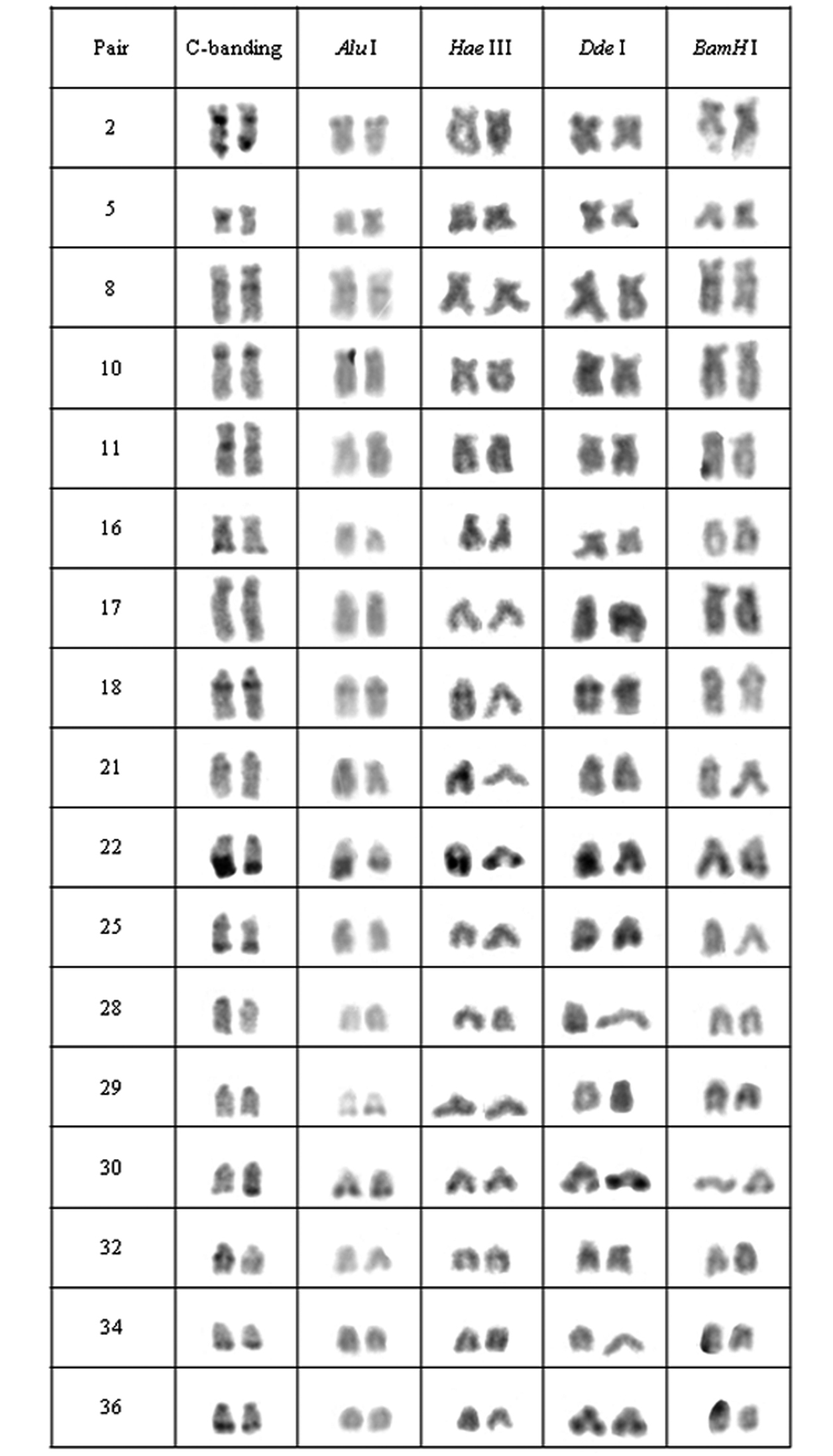
Chromosomal pairs from population B of Hypostomus prope unae showing the C-positive heterochromatin and banding pattern after digestion with the restriction endonucleases: Alu I, Hae III, Dde I and Bam HI.
Heteromorphic blocks were also evident in both populations. Besides the NOR-bearing pair, 18th, 21st and 37th pairs in population A and the 22nd pair in population B size differences between homologous (Figs 2, 3). Population C was characterized by interstitial and terminal marks in six chromosomal pairs, non-coincident with NORs (Fig. 4). On the other hand, population D presented eight pairs, most of them acrocentric, bearing interstitial C-bands and also interspersed with NORs (Fig. 5).
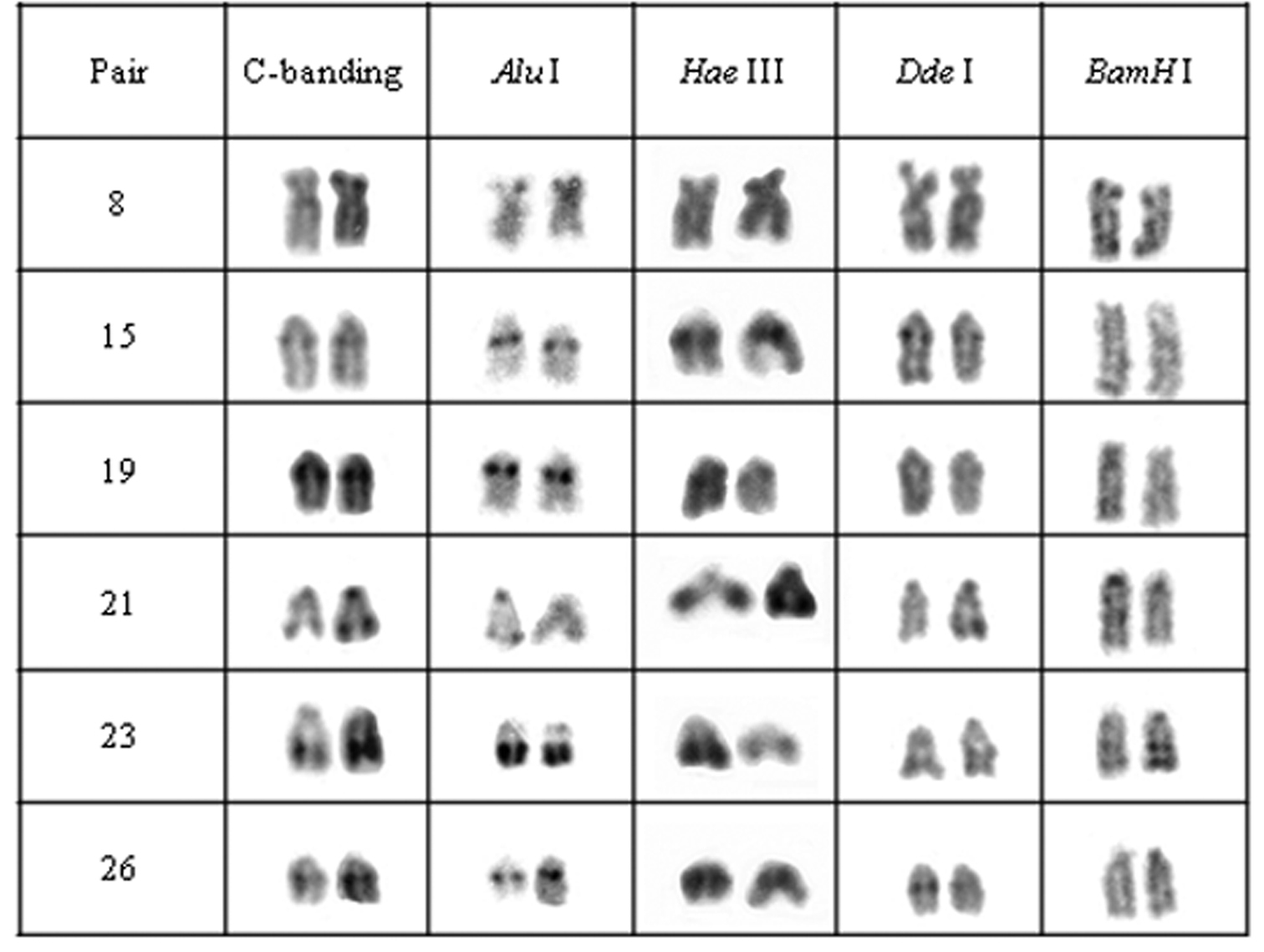
Chromosomal pairs from population C of Hypostomus prope unae showing the C-positive heterochromatin and banding pattern after digestion with the restriction endonucleases: Alu I, Hae III, Dde I and Bam HI.
Chromosomal pairs from population D of Hypostomus prope unae showing the C-positive heterochromatin and banding pattern after digestion with the restriction endonucleases: Alu I, Hae III, Dde I and Bam HI.
The digestion pattern using RE allowed identifying inter-population differences in several chromosomal regions but most in heterochromatin as shown in Table 2, where + stands for digested C-band and – stands for undigested heterochromatic region.
Heterochromatin digestion pattern using the restriction enzymes Alu I, Hae III, Dde Iand Bam HI per population of Hypostomus prope unae: (+) digested heterochromatin; (-) undigested heterochromatin; (±) partially digested heterochromatin.
| Population | C-banded pair. | Restriction Enzyme | |||
|---|---|---|---|---|---|
| AluI | IIIHae | IDde | HIBam | ||
| A | 1 | + | ± | + | + |
| 2 | + | + | + | + | |
| 3 | + | ± | + | + | |
| 7 | + | – | + | + | |
| 16 | – | – | – | – | |
| 17 | + | + | + | + | |
| 18 | – | – | – | – | |
| 21 | + | + | + | + | |
| 22 | + | – | + | + | |
| 23 | + | – | + | + | |
| 25 | – | – | – | – | |
| 28 | – | + | + | + | |
| 29 | ± | + | + | + | |
| 30 | + | + | + | + | |
| 32 | + | – | + | + | |
| 35 | + | ± | + | + | |
| 37 | + | – | – | – | |
| B | 2 | + | + | + | + |
| 5 | + | + | + | + | |
| 8 | – | + | + | – | |
| 10 | + | + | + | + | |
| 11 | + | + | + | + | |
| 16 | + | + | + | + | |
| 17 | + | + | + | + | |
| 18 | – | – | – | – | |
| 21 | + | + | + | – | |
| 22 | – | – | – | – | |
| 25 | + | + | – | + | |
| 28 | + | + | + | + | |
| 29 | + | + | + | + | |
| 30 | – | – | – | – | |
| 32 | + | – | + | – | |
| 34 | + | + | + | + | |
| 36 | + | + | – | + | |
| C | 8 | – | + | – | – |
| 15 | – | – | – | + | |
| 19 | – | – | + | + | |
| 21 | – | – | ± | ± | |
| 23 | – | – | – | ± | |
| 26 | – | – | – | + | |
| D | 2 | + | + | + | + |
| 4 | – | – | – | – | |
| 18 | – | – | – | – | |
| 22 | – | – | – | – | |
| 24 | – | – | – | – | |
| 27 | – | – | – | – | |
| 29 | – | – | – | + | |
| 33 | – | + | + | – | |
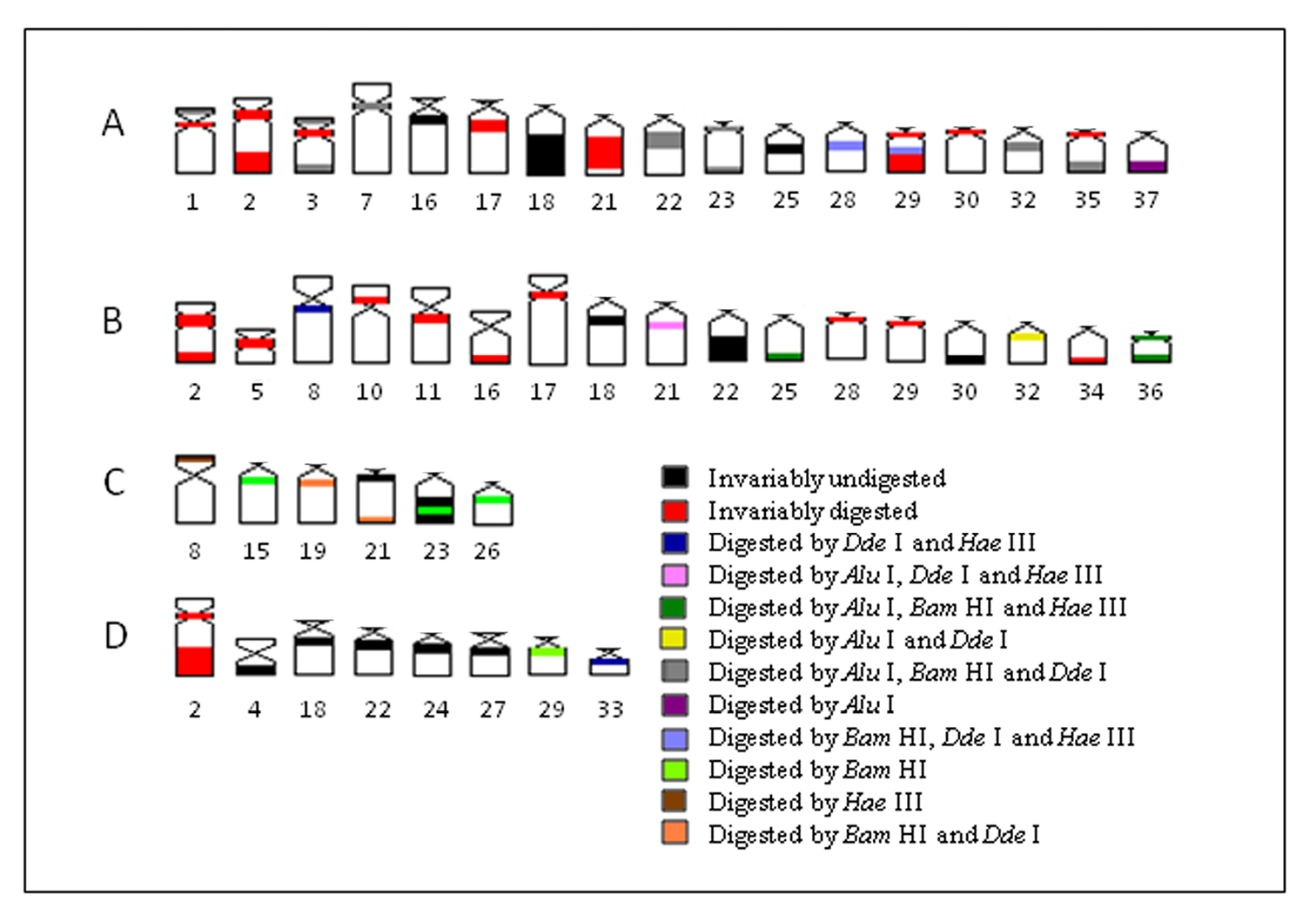
Five heterochromatin (or repeated DNA) groups were identified in population A: (a) the heterochromatin from pairs 2, 17, 21, 30, centromeric heterochromatin of pairs 1, 3, and 35, and terminal regions of the 29th pair were digested by all tested enzymes; (b) the chromosomal pairs 16, 18 and 25 lacked any target sequences; (c) pairs 7, 22, 23, 32 and the terminal heterochromatin of pairs 1, 3 and 35 were digested by Alu I, Bam HI and Dde I; (d) pair 28 and the upper portion of the heterochromatic block in pair 29 were digested by Hae III, Bam HI and Dde I; and (e) the 37th pair was digested by Alu I (Fig. 2).
In population B, the heterochromatin was divided into six groups: (a) the heterochromatin from pairs 2, 5, 10, 11, 16, 17, 28, 29 and 34 were digested by all enzymes; (b) the chromosomal pairs 18, 22 and 30 lacked the target sequences; (c) the pairs 25 and 36 were digested by Alu I, Bam HI and Hae III; (d) the heterochromatin from pair 8 was digested by Hae III and Dde I; (e) the 32nd pair was digested by Alu I and Dde I; (f) and the 21st pair was digested by Alu I, Hae III and Dde I (Fig. 3).
Enzymatic digestion of heterochromatic regions in population C revealed four heterochromatin groups: (a) centromeric region of pair 21 and the terminal blocks in pair 23 remained intact; (b) pair 8 was digested by Hae III; (c) pair 15, central portion of heterochromatic block in pairs 23 and were digested by Bam HI; (d) pair 19 and the terminal region of pair 21 were digested by Bam HI and Dde I (Fig. 4).
Heterochromatin regions in population D were also divided into four groups: (a) pair 2 was digested by all enzymes; (b) pairs 4, 18, 22, 24 and 27 were not digested by the tested enzymes; (c) the 29th pair presented target sequences for Bam HI; (d) and the 33rdpair was digested by Hae III and Dde I (Fig. 5).
Independently on the population, the nucleolus organizer regions (2nd pair) were digested by all restriction enzymes, including those samples in which NOR-associated heterochromatin was not detected by C-banding.
In relation to the digestion pattern in euchromatic regions, some conspicuous bands were observed, being specific for each population and enzyme. In general, population A presented a high number of chromosomes bearing Hae III bands, whereas populations B and C presented larger amounts of Alu I bands. On the other hand, population D was characterized by a large number of chromosomes bearing bands after treatments with all enzymes (data not shown).
DiscussionChromosomal digestion by restriction endonucleases results in a faint chromosomal staining and identification of a characteristic band pattern according to each enzyme (
A hindered access to chromosomal DNA has been pointed out as an alternative explanation for the banding profiles after RE digestion in some cases (
In the present work, the application of endonuclease treatments revealed a remarkable heterogeneity within heterochromatin among populations of Hypostomus prope unae, comprising either distinct or similar chromosomes, and even between heterochromatic segments. Based on these results, it was possible to identify inter- and intra-population (dis)similarities (Fig. 6). Most likely, the tested enzymes cleaved and removed DNA from both euchromatin and heterochromatin as demonstrated by some less stained chromosomal regions. Therefore, the observed bands can be regarded as non-removed DNA portions lacking the RE target sequences.
A–D Schematic ideogram of chromosomal pairs from populations A, B, C and D of Hypostomus prope unae, showing the combined banding pattern after digestion using Alu I, Bam HI, Hae III and Dde I.
The present data indicate that some heterochromatin regions in different chromosomes and/or populations share a similar composition, while others would present a unique composition. Thus, the banding pattern observed reflects directly the molecular nature of heterochromatin regions (
Such remarkable heterogeneous banding pattern shows that the populations of Hypostomus prope unae bear several heterochromatin families composed of distinct specific types of highly repetitive DNA. A similar finding was reported in the salmonids Salmo salar Linnaeus, 1758(
According to
Although inter-population differences were detected by both C-banding and RE digestion, some heterochromatin regions remained resistant to enzymatic digestion among populations, mainly in population D, revealing a higher differentiation in the DNA composition and/or heterochromatin organization in the latter. This population is also more divergent than the others because of its high frequency of interstitial C bands instead of terminal ones (Figs 5, 6).
Differences in heterochromatin patterns have been commonly reported in Neotropical fishes, including species from northeastern coastal basins (
It should be pointed out that the nucleolar organizer regions (2nd pair) was digested by all tested enzymes independently on the population, demonstrating that the distinct target sequences are “concertedly” interspersed along this region, even when NOR-associated heterochromatin was not detected, as observed in population C. Such behavior differs from the pattern observed by Sanches et al. (1990) that reported a differential NOR digestion indicative of a high amount of target sequences for Dde I and Hae III but a moderate number of restriction sites for Alu I.
Moreover, heteromorphic segments were observed between some chromosomal pairs in populations A (pairs 18, 21 and 37) and B (pair 22). Nonetheless, only the 21st pair in population A presented the target sequences for the selected RE, while the other heteromorphic segments proved to be resistant to their digestion activity.
Reports about restriction enzymes in Neotropical fish cytogenetics are scarce what hinders a detailed comparative analysis. However, this approach seems to be highly informative for species characterized by large amounts of heterochromatin as that presently studied, being able to reveal several genomic particularities. Moreover, repetitive DNA sequences might provide efficient chromosomal markers useful for evolutionary studies, identification of chromosomal rearrangements and sex differentiation (
As commonly reported in fishes of the genus Hypostomus (e.g.,
The authors would like to thank professors Ana Maria Waldschmidt (UESB) for providing the conditions and part of the material used in the present research, Cláudio Henrique Zawadzki (UEM-NUPELIA) for identifying the specimens, Antonio Carlos Bertollo (UFSCar) and André Laforga Vanzella (UEL) for their helpful comments and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for the financial support.