(C) 2011 Sandra Mariotto. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Populations of seven Ancistrus species were analyzed from streams and rivers of three hydrographic Brazilian basins. All populations showed different diploid numbers (2n), fundamental numbers (FNs), and karyotypes. Some representatives of Loricariidae have 2n = 54 chromosomes, which is very likely an ancestral cytotaxonomic characteristic, but many other representatives show extensive karyotype diversification. In the Ancistrus species studied, extensive karyotypic differentiation, which is generally associated with chromosome number reduction and rearrangement of the ribosomal RNA gene (rDNA) sites, was verified. Chromosomal locations of 18S and 5S rDNA were jointly detected using fluorescence in situ hybridization (FISH). In all the Ancistrus species analyzed, 18S rDNA sites were detected only on one chromosome pair, though this differed among species. 5S rDNA was located on 1–3 chromosome pairs either separately or in synteny with 18S rDNA in four of the seven species/populations. Hence the karyotype differentiation in Ancistrus species could be associated with a morphological speciation process, suggesting that chromosome fusions, inversions, deletions, duplications, and heterochromatination could contribute to the karyotype evolution of these neotropical armored catfishes.
karyotype evolution, Robertsonian rearrangement, Ag-NORs, 18S rDNA, 5S rDNA
In eukaryotes, 5S and 18S ribosomal genes (rDNA) are arranged into two distinct classes, namely the major rDNA family composed of 18S, 5.8S, and 28S genes and the minor family composed of 5S genes (
Studies that characterize the chromosomal locations of 5S and 18S rDNA in Siluriformes are scarce (
The classification of subfamilies within Loricariidae and the genera relationships have been targets of repeated reformulation (
Among Hypostominae, few species maintain the number of 54 chromosomes. All representatives in Ancistrini have 2n ≤ 54, indicating that centric fusions contributed to the karyoevolution of this tribe. In this study, we used comparative chromosomal markers to establish chromosome homologies among some Ancistrus species and investigated the cytotaxonomical, biogeographical, and karyoevolutionary features of this group.
Materials and methodsOne hundred and thirty six specimens [male (M) and female (F)] of seven Ancistrus species were cytogenetically analyzed. All specimens were from rivers and streams of three hydrographic basins (a, b, and c) of Mato Grosso state, Brazil: (a) Paraguay basin, Coxipó river, 15°21'59"S, 55°57'11"W, Ancistrus claro Knaack, 1999 (11 M and 10 F);Sepotuba river, 14°41'35"S, 57°48'14"W, Ancistrus sp. 04 (12 M and 15 F); Currupira river, 15°07'59"S, 56°49'47"W, Ancistrus sp. 08 (7 M and 8 F); Flechas stream, 15°58'7"S, 57°19'7"W, Fundo stream, 16°14'17"S, 56°37'31"W, and Pari stream, 15°36'6"S, 56°12'19"W, Ancistrus cf. dubius Eigenmann and Eigenmann, 1889 (2 M and 2 F from each locality); Arrombado bay, 16°21'21"S, 56°27'55"W, Ancistrus cuiabae Knaack, 1999(15 M and 15 F); (b) Araguaia–Tocantins basin, Salgadinho stream, 14°40'14"S, 52°21'50"W, Ancistrus sp. 13 (11 M and 6 F); and (c) Amazon basin, Matrixã river, 10°3'7"S, 57°36'27"W, Ancistrus sp. 06 (9 M and 5 F).
Specimens were morphologically identified and deposited in the Museu de Ciências da Pontíficia Universidade Católica do Rio Grande do Sul (MCP/PUC; MCP 41966, 41968, 41971, 41973, 41975, 41978, 41979) and Núcleo de Pesquisas Limnológicas da Universidade Estadual de Maringá, Paraná (NUPELIA/UEM; NUP 6827, 7492). Ancistrus sp. 04, 06, 08, and 13 present discriminative morphological characteristics that have not yet been described.
Chromosomal preparations were obtained from anterior kidney cells using an in vivo treatment with colchicine (
FISH was performed according to
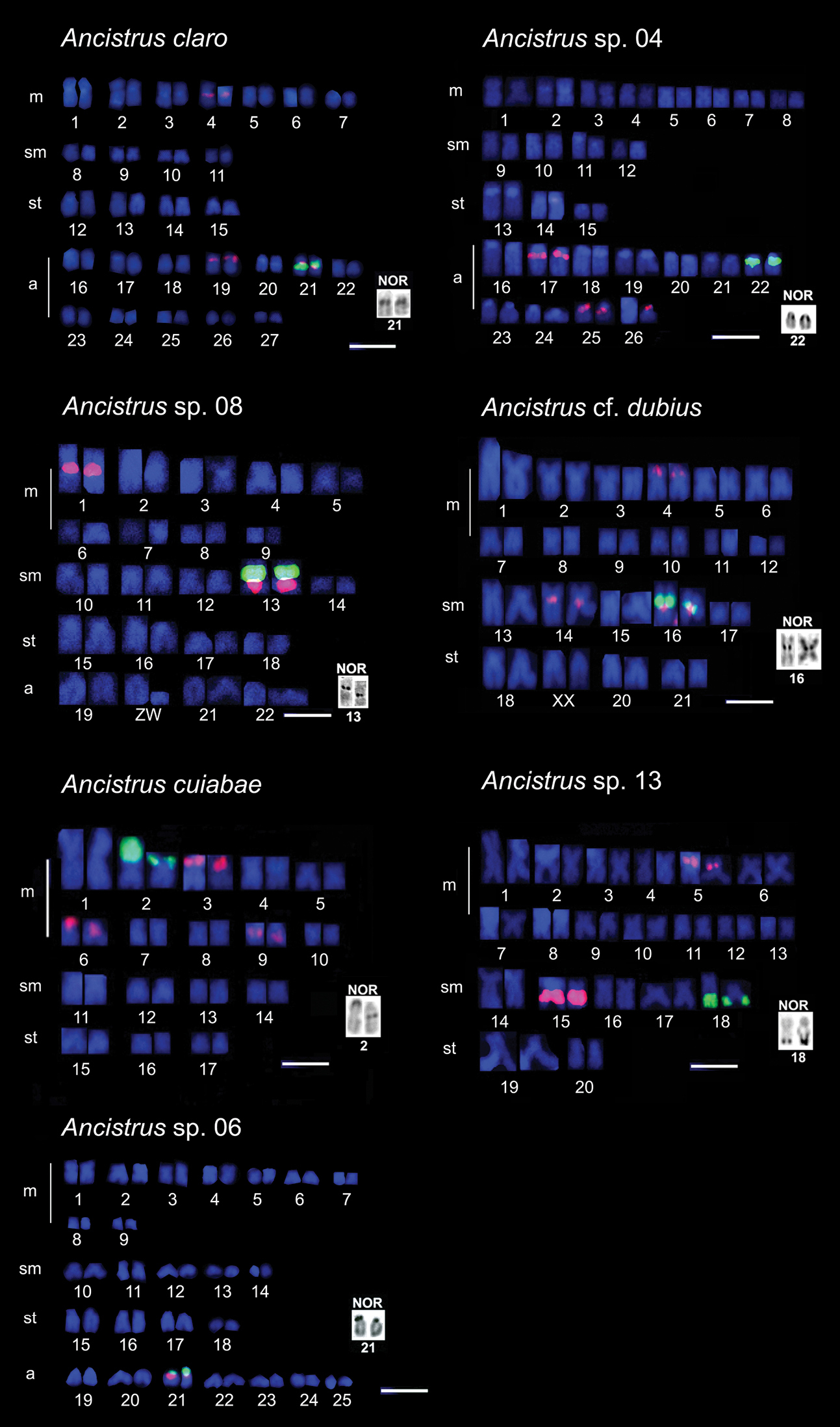
The studied species showed variations in 2n and in karyotypic formulae (Table 1, Fig. 1). The 2n ranged from 54 chromosomes in Ancistrus claro to 34 chromosomes in Ancistrus cuiabae. The fundamental number (FN) varied from 68 to 86 chromosome arms (Table 1). NORs were seen in a single chromosome pair in all the Ancistrus sp. analyzed using silver nitrate staining and FISH with 18S rDNA probe (Fig. 1). However, an interspecific variation was observed in the NOR-bearing chromosome pairs and NOR locations in these chromosomes (Table 1, Fig. 1 and 2).
Chromosomal data in analyzed Ancistrus species.
| Species/basin | 2n | Karyotypic formulae | FN | SC | rDNA synteny |
|---|---|---|---|---|---|
| Paraguay basin | |||||
| Ancistrus claro | 54 | 14m+8sm+8st+24a | 84 | - | Present |
| Ancistrus sp. 04 | 52 | 16m+8sm+6st+22a | 82 | - | Absent |
| Ancistrus sp. 08 | 44 | 18m+10sm+8st+8a | 80 | ZZ/ZW | Present |
| Ancistrus cf. dubius | 42 | 24m+10sm+8st | 84 | XX/XY | Present |
| Ancistrus cuiabae | 34 | 20m+8sm+6st | 68 | - | Absent |
| Araguaia–Tocantins basin | |||||
| Ancistrus sp. 13 | 40 | 26 m+10sm+4st | 80 | - | Absent |
| Amazon basin | |||||
| Ancistrus sp. 06 | 50 | 18m+10sm+8st+14a | 86 | - | Present |
Chromosomes of Ancistrus species after dual color-FISH showing 5S rDNA (red) and 18S rDNA (green) sites. Silver nitrate-stained nucleolar organizing region (Ag-NOR) patterns are shown in the boxes. Bars = 10 µm.
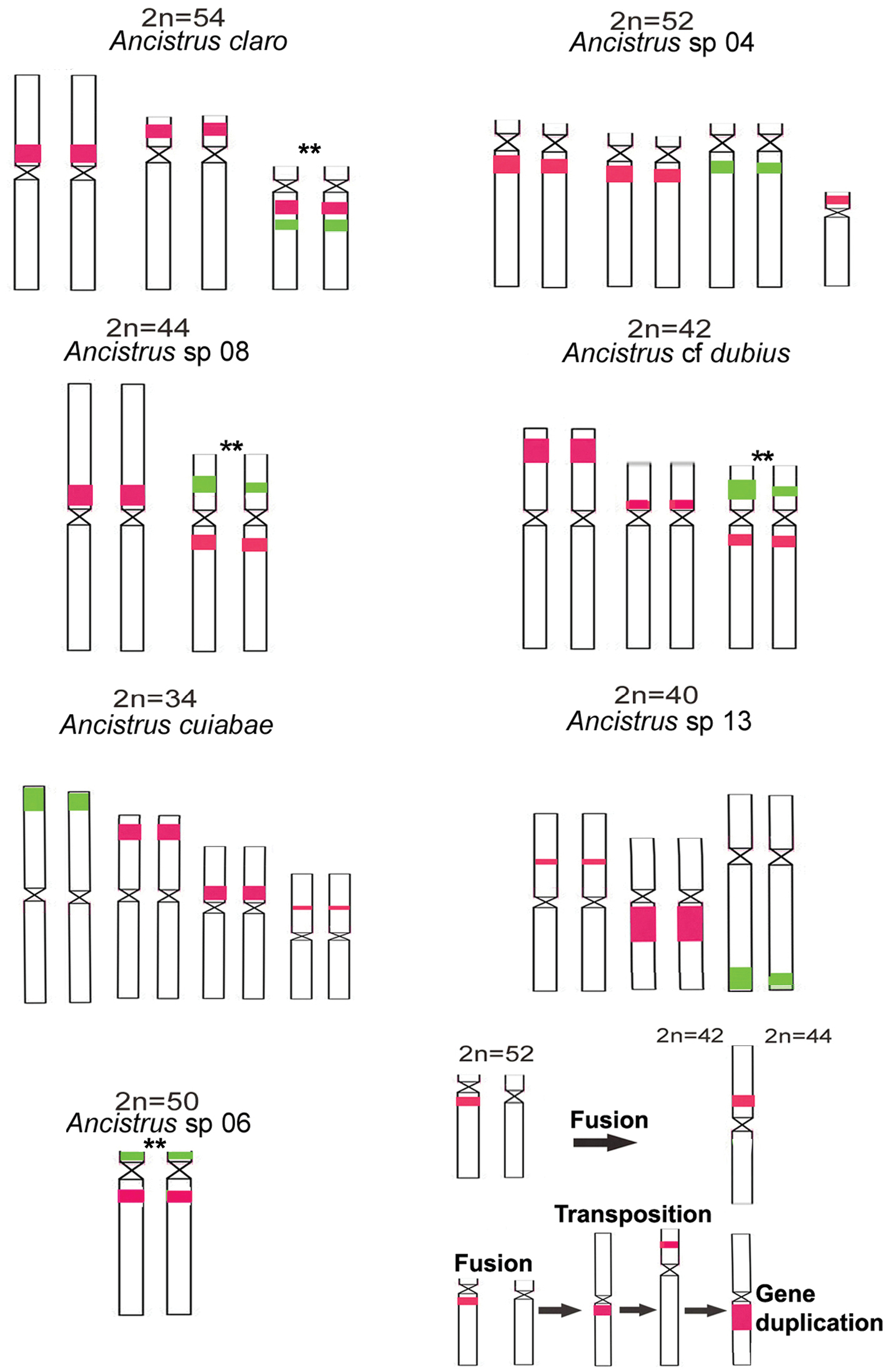
Idiograms of chromosomes bearing 5S (red) and 18S (green) rDNA (a–g); (h) probable chromosomal rearrangements (fusions, transpositions, and gene duplication) occurred during the evolution of Ancistrus species; ** denotes possible homeologous chromosomes.
Diploid number (2n), metacentric (m), submetacentric (sm), subtelocentric (st), acrocentric (a), fundamental number (FN), sex chromosome system (SC).
The number of 5S rDNA sites varied among species (Fig. 1). Multiple locations were observed in all species, except Ancistrus sp. 06 (Fig. 1). Dual-color FISH using 5S and 18S probes showed co-localized sites on apparently homeologous chromosome pairs in Ancistrus claro, Ancistrus sp. 08, Ancistrus cf. dubius, and Ancistrus sp. 06 (Fig. 1). However, in Ancistrus sp. 04, Ancistrus cuiabae, and Ancistrus sp. 13, 18S and 5S rDNA probing revealed different chromosome pairs carrying 18S and 5S rDNA (Fig. 1). Ancistrus claro showed syntenic rDNA site in pair 21 and four additional 5S rDNA sites (Fig. 1). In Ancistrus sp. 04, syntenic 18S and 5S rDNA classes in pair 22 and additional 5S rDNA sites in pairs 17 and 25, as well as one homologue in pair 26 were visualized (Fig. 1). Ancistrus sp. 08 showed 18S rDNA syntenic to 5S rDNA in pair 13 and an additional 5S rDNA site in pair 1 (Fig. 1). In addition, Ancistrus sp. 08 had a heteromorphic ZZ/ZW sex chromosome system in pair 20 (Fig. 1). Ancistrus cf. dubius showed syntenic 18S and 5S rDNA classes in pair 16, and additional 5S rDNA sites in pairs 4 and 14 (Fig. 1). This species presented an extensive heterochromatic region in pair 19 of females and in one of the complements of pair 19 of males, thus characterizing a sexual chromosomal system, XX/XY (data not shown). Ancistrus cuiabae showed 18S rDNA located in pair 2 and three different pairs (3, 6, and 9) carrying 5S rDNA sites (Fig. 1). In Ancistrus sp. 13, 18S rDNA was located in pair 18, and 5S rDNA was located in pairs 5 and 15 (Fig. 1). Ancistrus sp. 06 showed syntenic rDNA classes in pair 21 (Fig. 1).
DiscussionThe catfish Loricariidae is one of the most speciose components of neotropical freshwater fish fauna. The karyotypic differentiation of Ancistrini is correlated with the great diversification of forms in this tribe and may play an important role in the genetic/reproductive isolation of species (
This study revealed that chromosome fusion is the major mechanism in 2n reduction of some Ancistrini species. In the sister group Hypostomini, in which all species present 2n ≥ 54, it has been postulated that the increase in the subtelocentric/acrocentric chromosome number is directly proportional to 2n, thereby indicating that centric fissions have played a key role in karyotype evolution of the group (
Ag-NORs can also be used as efficient markers in Loricariidae. A single NOR pair in an interstitial location is considered a primitive characteristic in Loricariidae and is maintained in most Ancistrini species (
Ancistrus claro, Ancistrus cf. dubius, Ancistrus sp. 08, and Ancistrus sp. 06 conserve the interstitial NORs in a putative homologous pair. However, the chromosomal morphologies of NOR-bearing chromosomes vary, probably because of the accumulation of adjacent heterochromatin (
FISH mapping of 5S rDNA in Ancistrus species showed variations in the number and shape of chromosomes bearing this ribosomal family. Most sites were observed in the interstitial portion of the long or short arm of chromosomes; however, in some cases, such as Ancistrus cf. dubius and Ancistrus sp. 08, pericentromeric 5S rDNA sites were visualized. The occurrence of multiple and variable 5S rDNA can be considered an important process underlying this huge karyotypic diversity. In the subfamily Loricariinae,
Based on the trends in the karyotype evolution in Ancistrini, it can be inferred that the variation in 2n (54–34 chromosomes) in the Ancistrus species studied could possibly involve several chromosomal rearrangements and gene flow restriction in different hydrographic basins or rivers (Fig. 2). The 2n primitive was found in Ancistrus claro from the Paraguay basin, which presents syntenic 18S and 5S rDNA sites in pair 21 and no sex chromosome heteromorphism. However, this species presents two additional 5S rDNA sites, a characteristic considered apomorphic in this family. Ancistrus sp. 04 from the Paraguay basin presents 2n = 52 chromosomes, no syntenic rDNA sites, no sex chromosome heteromorphism, and additional 5S rDNA sites. Hence, rDNA translocation and fusion chromosomes could have occurred in species diversification, and the broken condition of the syntenic rDNAs could have originated in one lineage with Ancistrus cuiabae and Ancistrus sp. 13. The similarity among them is maintained by no syntenic rDNA sites, no sex chromosome heteromorphism, and closely hydrographic basins. However, the pair carrying 18S rDNA in Ancistrus sp. 04 is apparently homeologous to chromosome pair 18 in Ancistrus sp. 13, which could have originated in the second pair in Ancistrus cuiabae by chromosome fusion.
The other lineage consists of species that retain the syntenic rDNA sites (Ancistrus claro, Ancistrus sp. 08, Ancistrus cf. dubius, and Ancistrus sp. 06). In addition to Ancistrus claro, species from the Amazon basin (Ancistrus sp. 06) have 2n = 50 chromosomes, considered to have been derived from the family. However, this species retains a primitive single NOR pair with syntenic 5S rDNA site and no additional site. Thus, the chromosome number reduction in Ancistrus sp. 06 is attributable to chromosome fusion. Ancistrus sp. 08 and Ancistrus dubius from the Paraguay basin have chromosome number reduction (2n = 44 and 2n = 42, respectively) by fusion and independent pathways to differentiated sex chromosome systems (
Nevertheless, there is large chromosome plasticity among the species from the Paraguay basin, and the diversity of chromosome types with 5S and 18S ribosomal cistrons in Ancistrus sp. explain the high degree of karyotypic diversification in this taxon. Also, the 18S rDNA marker, which is mostly considered to be conserved in a single chromosome pair in the interstitial position, showed different site locations in different types of chromosomes.
The variation observed in the 2n, FN, and rDNA sites of the Ancistrus sp. could be attributed to structural and numeric chromosome rearrangements. The karyotypic data presented here are important tools for taxonomy of Ancistrus species. The karyotype differentiation in Ancistrini could be associated with a morphological speciation process, suggesting that chromosome fusions, inversions, deletions, duplications, and heterochromatination could contribute to the chromosomal differentiation of Ancistrini.
The authors are grateful to Alexandre Cardoso, Tiago Carvalho (MCT/PUC/RS) and Claudio Zawadzki (NUPELIA/UEM) for the identification of this species. This work was partially supported by FAPEMAT (Fundação de Amparo a Pesquisa de Mato Grosso) CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and the Fundação Araucária (Fundação Araucária de Apoio ao Desenvolvimento Científico e Tecnológico do Estado do Paraná).