(C) 2011 Marlykynti Hynniewta. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Abstract
Ten Citrus (Linnaeus, 1753) species of North-East India have been karyo-morphologically analysed. All studied species had 2n=18 chromosomes without any evidence of numerical variation. All the chromosomes were found to be of metacentric and sub-metacentric in all the species; the morphology of the chromosomes showing size difference only. Symmetrical karyotype which does not have much difference in the ratio of longest to shortest chromosome in all the species was observed. Three species, Citrus grandis (Osbeck, 1757), Citrus reticulata (Blanco, 1837) and Citrus medica (Linnaeus, 1753) are identified as true basic species from asymmetry studies of karyotypes as they reflect on the primitive nature of their genomes. Citrus indica (Tanaka, 1937)occupies a special taxonomic position within the genus Citrus as a progenitor for other cultivated species.
Citrus, karyotype, genetic variability, asymmetry index
The genus Citrus is economically very important and is known for its juice and pulp throughout the world. The genus belongs to the family Rutaceae that includes 162 species (
The south-east Asia, Australia and the intervening island-areas between Australasia and Central Africa and the north-eastern region of India along with neighbouring China (Mc Phee 1967,
South and western hills of Meghalaya in the North-East are reported to have maximum diversity for Citrus reticulata, Citrus grandis, Citrus limon and Citrus aurantifolia. These are extensively cultivated for their taste, good pulp and have very high market demand. Citrus indica is supposed to be the most primitive species and perhaps the progenitor of cultivated Citrus (
The relationship between the species within the genus Citrus has been made complicated due to combination of factors such as wide cross compatibility, repeated cross pollination and apomixis. Wide hybridization in Citrus affects karyotype stability (
The cytogenetical characterization of Citrus accession could help in the identification of a particular genomic variant, or for the detection of true hybrids in breeding program, as well as for studies of karyotypes evolution of the group (
The plant material used in the present investigation was collected from various region of North-East India and the vouchers specimens have been submitted to National Herbarium of Crop Plants, National Bureau of plant Genetics Resources, New Delhi (Table 1). The plants were grown in green house of Plant Biotechnology Laboratory, Department of Biotechnology and Bioinformatics of North-Eastern Hill University, Shillong. For each species, wherever possible, a minimum of five individuals and more than one population were analyzed. For obtaining actively growing root tips, plants were raised in earthen pots and the root tips of about (0.5–1.0 cm) long were excised. All the root tips were pre-treated with 8-hydroxyquinoline (0.002M) for three hours at room temperature, fixed in ethanol-acetic acid (v/v, 3:1) and subsequently stored at 4 oC until required. For slide preparation, the root tips were washed twice in distilled water, hydrolysed in 5N HCl for 20 min at room temperature. The hydrolysed root tips were washed in distilled water and stained in Feulgen stain for 45 min. The root tips were subsequently squashed in 1% acetocarmine. The micro-photographs were taken using Jenoptik CCD camera (Germany) attached to labomed LX 400 brightfield microscope. At least five clear preparations of chromosome complements of each species were analyzed for the karyotypes. Idiograms were prepared from photo-micrographs by cutting out individual chromosomes, arranging them in descending order of their length and matching on the basis of morphology. The standard method of chromosome classification (
Citrus species used in the present investigation.
| Sl. No. | Species | Common Name | Collection No. | Source |
|---|---|---|---|---|
| Subgenus Citrus | ||||
| 1 | Citrus reticulata | Khasi Mandrin | CR-9 | Pynursla |
| 2 | Citrus jambhiri | Rough lemon | CJ-6 | Wahkhen |
| 3 | Citrus sinensis | Sweet orange | CS-2 | Shillong |
| 4 | Citrus limon | Assam Lemon | MD/33 | Mizoram |
| 5 | Citrus grandis | Pummelo | CG-7 | Ri Bhoi |
| 6 | Citrus limetta | Sweet limes | CLe-1 | Shillong |
| 7 | Citrus indica | Indian wild orange | SO1 | Nokrek, Garo hills |
| 8 | Citrus medica | Citron | CMi-2 | Wahkhen |
| Subgenus Papeda | ||||
| 9 | Citrus macroptera | Melanesian Papeda | CMa-1 | Cherrapunjee |
| 10 | Citrus latipes | Khasi Papeda | Clt-2 | Upper Shillong |
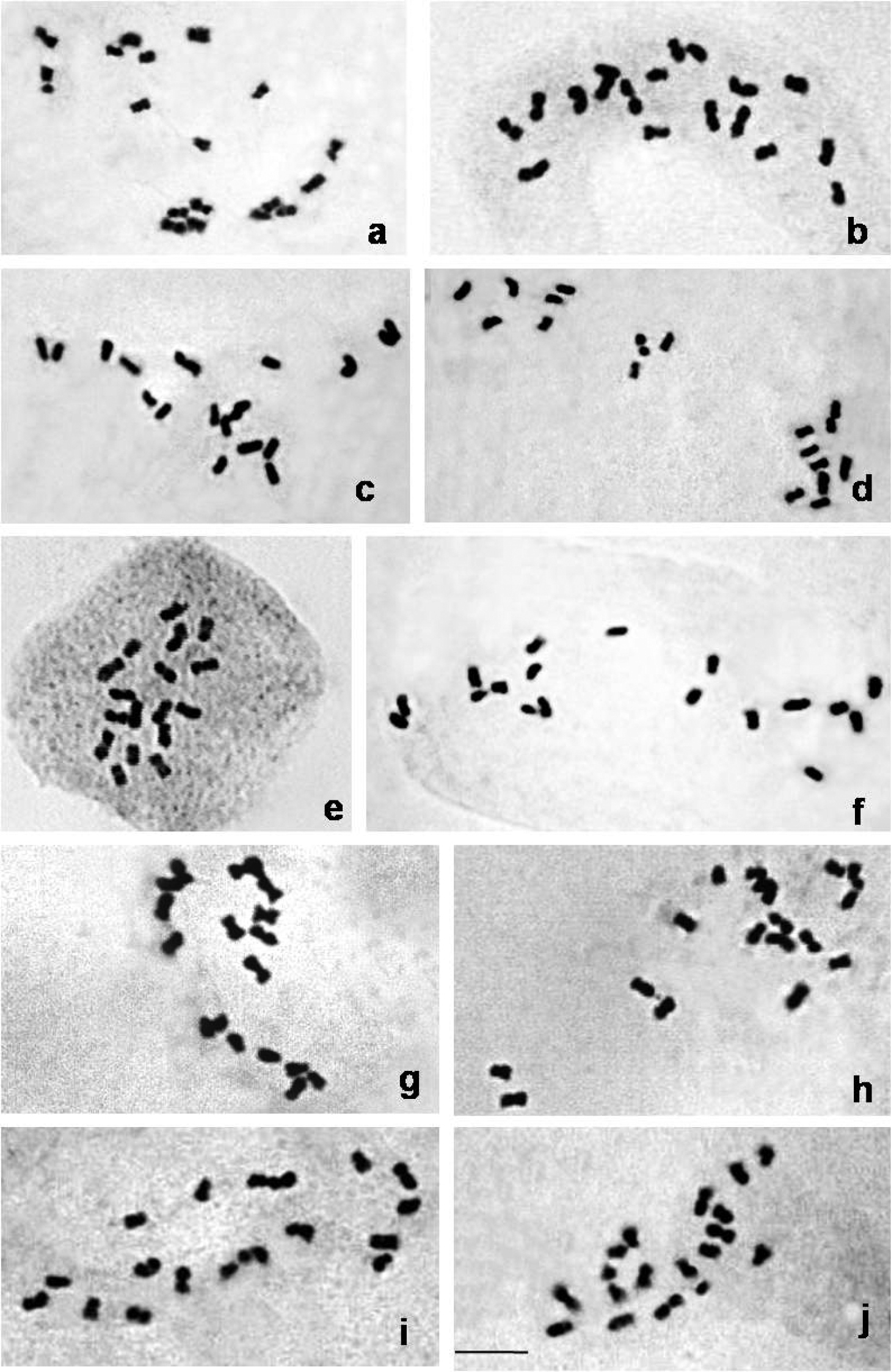
The data related to chromosome complements/karyotypes have been presented in Table 2 and illustrated in Fig. 1 and 2 and it is amply clear that among the 10 species of Citrus presently studied, two species namely Citrus jambhiri Lushington, 1910 and Citrus limon Linnaeus, 1753 were characteristic in having exclusively sub-metacentric chromosomes in the chromosomes complements. On the other hand the remaining 8 species namely Citrus macroptera, Citrus grandis, Citrus medica Linnaeus, 1753, Citrus reticulata, Citrus sinensis, Citrus latipes, Citrus indica and Citrus limetta Linnaeus, 1753 had at least one pair of metacentric chromosome among the chromosome complements. It was more intriguing to record that two metacentric pairs were observed in Citrus reticulata and Citrus latipes as metacentrics while one pair of metacentric were recorded in remaining 6 species. Further the position of the meta-centrics varied in different species of Citrus presently studied ranging from 2nd pair (in Citrus grandis and Citrus latipes), 3rd pair (in Citrusreticulata), 4th pair (in Citrus macroptera and Citrus indica), 5th pair (in Citrusreticulata, Citrus latpipes and Citrus limetta), 7th pair (in Citrus sinensis) and 8th pair (in Citrus medica). Thus, the 6th and 9th pairs in all the species have been found to be invariably sub-metacentric.
Karyotype formulae and characteristics in 10 species of Citrus. AI- asymmetry index; SC - the shortest chromosome length; LC - the longest chromosome length; CL - mean length of chromosome; CI - mean centromeric index; SD - standard deviation; CVCL- component expressing the relative variation in chromosome length; CVCI - component expressing the relative variation in centromeric index.
| Species | Collection No | 2n | Number of second-dary constriction | Range SC-LC (μm) | Ratio LC/SC |
CL (μm) Mean (±SD) |
CI Mean (±SD) |
CVCL | CVCI | AI | Karyotype formula* |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Citrus macroptera | Cma-1 | 18 | - | 5.01–10.52 | 2.09 | 7.44 (±1.9) |
40.99 (±5.4) |
25.53 | 13.17 | 3.36 | 16L+2V |
| Citrusgrandis | CG-7 | 18 | 2 | 4.03–11.12 | 2.75 | 7.83 (±2.04) |
43.93 (±3.2) |
26.05 | 7.28 | 1.89 | 16L+2V |
| Citrusmedica | Cmi-1 | 18 | - | 4.51–12.02 | 2.66 | 6.88 (±1.84) |
42.73 (±3.0) |
26.74 | 7.02 | 1.87 | 16L+2V |
| Citrusreticulata | CR-9 | 18 | - | 4.01–9.03 | 2.25 | 6.51 (±1.5) |
43.07 (±4.6) |
23.04 | 10.68 | 2.46 | 14L+4V |
| Citrus sinensis | CS-2 | 18 | - | 4.03–9.51 | 2.35 | 6.71 (±1.34) |
43.88 (±6.7) |
19.97 | 15.26 | 3.04 | 16L+2V |
| Citrus jambhiri | CJ-6 | 18 | - | 4.04–9.02 | 2.23 | 5.51 (±1.2) |
38.72 (±6.7) |
21.77 | 17.3 | 3.76 | 18L |
| Citrus latipes | CLt-1 | 18 | - | 4.02–10.11 | 2.51 | 7.08 (±1.76) |
39.97 (±6.9) |
24.85 | 17.26 | 4.28 | 14L+4V |
| Citrus indica | SO1 | 18 | - | 4.01–8.14 | 2.02 | 5.81 (±1.28) |
43.1 (±3.8) |
22.03 | 8.81 | 1.94 | 16L+2V |
| Citrus limon | MD/33 | 18 | 2 | 4.03–9.10 | 2.25 | 6.38 (±1.39) |
42.16 (±5) |
21.78 | 11.85 | 2.58 | 18L |
| Citrus limetta | Cle-1 | 18 | - | 3.51–9.01 | 2.56 | 6.18 (±1.77) |
42.48 (±4.8) |
28.64 | 11.29 | 3.23 | 16L+2V |
* As per the method of
Mitotic complements of 10 Citrus species (2n=2x=18). a Citrus macroptera, b Citrusgrandis, c Citrus medica, d Citrus reticulata, e Citrus sinensis, f Citrus jambhiri, g Citrus latipes, h Citrus indica, i Citrus limon, j Citrus limetta. Bar = 5µm.
Karyograms of 10 Citrus species. a Citrus macroptera, b Citrus grandis, c Citrus medica, d Citrus reticulata, e Citrus sinensis, f Citrus jambhiri, g Citrus latipes, h Citrus indica, i Citrus limon, j Citrus limetta. Bar represent heteromorphic pairs.
Sub-telocentric and telocentric chromosomes which are presumed to significantly influence the symmetry of the karyotype were alltogether absent in any of the species presently studied. From the details of karyotypic formula derived for various species of Citrus, three patterns of karyotype formulae, 18L, 16L+2V and 14L+4V, were recorded. The ratio of longest to shortest chromosomes was recorded as highest in Citrus grandis and the lowest in Citrus indica.
Partial homology among the somatic chromosomes is often expressed in the form of heteromorphism and heteromorphic pairs in karyotypes. The present observation of 10 different species of Citrus had shown interspecific diversity with regards to presence or absence of heteromorphic pair in the chromosome complements. Citrus macroptera, Citrus reticulata, Citrus limon and Citrus latipes were characteristic in lacking any heteromorphic pair, while Citrus grandis, Citrus medica and Citrus limetta are unique in having two pairs of heteromorphic chromosomes in their respective complements. One pair of heteromorphic chromosomes was characteristic in Citrus sinensis, Citrus jambhiri and Citrus indica.
Due to technical problems nucleolar chromosome could not be clearly scored in any of the species presently studied, although there were some indications to suggest that the second pair in Citrus grandis and third pair in Citrus limon are probably nucleolar in nature by revealing the secondary constriction.
The asymmetry index (AI) value which has been derived from the data related to Chromosome length (CL) and Centromeric index (along with the co-efficient of variation) has resolved the ten species of Citrus presently investigated into two groups, one with low value of asymmetry index indicating high karyotype symmetry corresponding to Citrus medica (1.87), Citrus grandis (1.89), Citrus indica (1.94), Citrus reticulata (2.46). The other group with high asymmetry index indicate low karyotype symmetry corresponding to Citrus sinensis (3.04), Citrus limetta (3.23), Citrus macroptera (3.36), Citrus jambhiri (3.76) and Citrus latipes (4.28). Citrus limon reported to be an intermediate species had an asymmetry index value of 2.58 indicating its link between the above two groups.
DiscussionFrom the perusal of published literature it can be seen that the somatic chromosome number in the genus Citrus is diverse ranging from 2n=18, 27, 36, 54, etc. (
Thus the present data as reflected from Fig. 1 and 2, combined with chromosome counts available from the literature confirms that the genus Citrus is apparently monobasic in nature and x=9 is the most acceptable number. Such observation received an ample support from reports of
In the present studies on 10 different Citrus species, the chromosome complements were all resolved into either metacentric or sub-metacentric chromosomes only. From the details of karyotypic formulae derived for these species of Citrus, three patterns of karyotype formulae, 18L, 16L+2V and 14L+4V, were recorded and there was complete absence of sub-telocentric and telocentric chromosomes which is indicative of the stability of the genome and of the absence of structural alteration of the chromosomes in the genus Citrus. Therefore, it is presumed that speciation in the genus Citrus could have been influenced by gene mutations which have no effect in the overall structure of chromosomes.
From the karyological data presented in Table 2 it can be observed that the asymmetry index of different species of Citrus presently investigated had shown significant variation. Citrus medica, Citrus grandis and Citrus reticulata which are considered as true basic species (
The present work was carried out in the Department of Biotechnology and Bioinformatics, North-Eastern Hill University, Shillong and was supported by University Grants Commission, New Delhi through scholarship.