(C) 2011 L.A. Lavrenchenko. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Three new karyotypes (2n=40, 44, 52) are described revealing what are probably new cryptic species of Ethiopian spiny mice. Two other diploid numbers have already been reported for the country (2n=36 and 68) and, overall, the five known karyotypic forms constitute a common lineage differentiated by a Robertsonian process. Such arrays of karyotypic forms are known as a ‘Robertsonian fan’. This view of the situation in Ethiopian Acomys I. Geoffroy, 1838 is based on standard chromosomal morphology that reveals a constant FN (68) and needs further investigation of chromosome homology by differential staining and/or molecular cytogenetic techniques as well as further molecular phylogenetic analysis.
mammalian karyotype, Robertsonian variation, Acomys
Karyotypic studies on mammals of Ethiopia play an important role in species identification and in the interpretation of phylogenies of taxonomic groups (
It became clear from early descriptions of the Middle East populations that a special type of chromosomal variation, known as Robertsonian translocations, is to be regarded as the principal way of karyotypic evolution both between and within Acomys taxa. The differences in diploid numbers that was defined initially in representatives of Acomys cahirinus (É. Geoffroy, 1803) and Acomys russatus (Wagner, 1840) from Israel (2n=38 and 66, respectively), have shown almost exactly the 2n limits in the whole genus (
The variation in chromosome arm numbers (the Fundamental Number, otherwise FN) is relatively low, from 66 to 78 (
In Ethiopia, only karyotypes with the extremal 2n values were previously found which have been reported for the two species inhabited the Lower Omo River Valley in the very south (2n=36 in Acomys percivali Dollman, 1911 and 2n=60 in Acomys wilsoni Thomas, 1892, according to
In this communication, three new karyotypes of Acomys are presented which adds substantially to the 2n range known for taxa inhabiting this country (40, 44 and 52 versus 36 and 60, 68) and thus extends the series of Robertsonian transformations, known as a ‘Robertsonian fan’, performed in the related karyotypes. This study covers also three new geographic parts of Ethiopia where the spiny mice were never examined.
Materials and methodsAnimals were collected during the 2008 and 2010 field seasons in the course of the Joint EthioRussian Biological Expedition (JERBE). Chromosome preparations were obtained from five Acomys specimens collected in two lowland regions on opposite sides of the Rift Valley. Among them, two specimens of both sexes were from a geographic site in the east (Babille Elephant Sanctuary, 9.0601°N; 42.2699°E, 1200 m a.s.l.) and three specimens from two sites of the Alatish National Park, north-western Ethiopia (Amjale, 12.3875°N; 35.7309°E, 533 m a.s.l., 2 males, and Bermil, 12.4958°N; 35.6382°E, 575 m a.s.l., 1 female).
For the karyotype analysis, bone marrow cell suspensions were prepared according to the standard colchicine, hypotonic solution and air-dried technique (
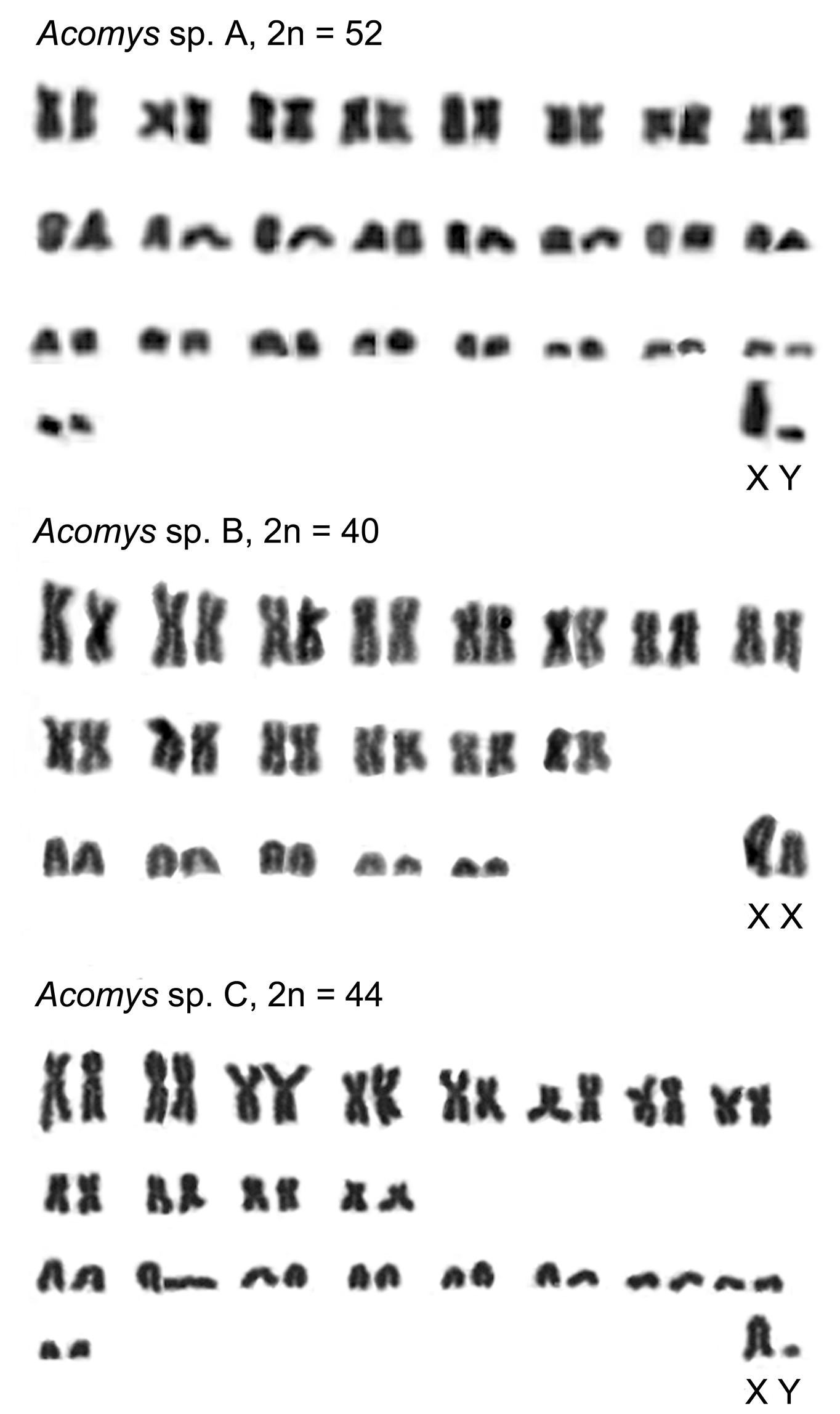
Three different karyotypes are found which characterize the Ethiopian Acomys of lowland savanna from three geographic samples in two different parts of the country. A higher chromosome number (2n=52) is detected in a male from Amjale. Its chromosome set contains lesser number of metacentric pairs than acrocentric ones (8 and 18, respectively; FN=68). Sex chromosomes cannot be reliably identified without differential staining, however, a larger acrocentric element with a distinguished short arm was recognized as the X chromosome, when one of the smaller acrocentrics may represent the Y chromosome. The autosomal arm number (FNa) characteristic for this karyotype is 66.
The lesser chromosome numbers are found in two other karyotypes (Figure 1). 2n=44 is characteristic for animals from the Babille Elephant Sanctuary. In this karyotype 12 metacentric and 10 acrocentric pairs are present (FN=68). The acrocentric chromosomes apparently include the XX pair in females and XY in males. NFa is 66 in this case. 2n=40 is reported from Bermil, and only 6 pairs of acrocentrics are identified in this karyotype (FN=68), one of which may be determined as the XX pair in a female studied, by analogy with the sex chromosomes of other Acomys species (Table 1). NFa is again 66.
Karyograms of Ethiopian Acomys spp. A – Amjale, B – Bermil, C - Babille.
Description of three new karyotypes of Acomys presented in this study indicates that there is more karyotype diversity in Ethiopian Acomys than it was thought previously. Three additional 2n numbers that were found (40, 44, and 52) fill in the series of 2nchanges where only minimal (36) and maximal (60, 68) limits were known until now (Table 1).
The common feature for five out of six karyotypes is the same number of chromosome arms (FN). The FN value is invariably 68 for full chromosome complements and 66 for the autosomes (FNa) in the karyotypes with various 2n which were attributed to at least five species, including Acomys cahirinus (2n=36) and four unidentified taxa, probably cryptic species - Acomys sp. (2n=68), Acomys sp. A (2n=52), Acomys sp. B (2n=40), and Acomys sp. C (2n=44) (
Regarding this Ethiopian group, interrelations based on Robertsonian rearrangements could be suggested. It is well known that it is usually difficult to decide whether Robertsonian fusions or fissions occurred in each given group, and the interpretation of karyotype evolution depends upon the context of the comparative analysis. It is often a working hypothesis that a chromosome set with the most numbers of acrocentrics is the primitive one (2n=68 in the case of Acomys) and that karyotypic evolution should pass via successive fusions of any twin acrocentrics into one metacentric. In fact, due to the bi-directional Robertsonian process and probable multiple fusions of original acrocentrics, the chromosomal sibling species may exist that should confuse a hypothetic common chain of karyotypic changes from maximal to minimal 2n, or, in our case, from 2n=68 to 2n=36 via 2ns such as 52, 44 and 40. Indeed, a clear indication on Robertsonian, or centric fission has been obtained through the karyotype comparison between the continental (Turkey) Acomys cilicicus Spitzenberger, 1978 and Mediterranean sea island (Cyprus) spiny mouse, Acomys nesiotes (Bate, 1903), which differ in 2n as 36 and 38, respectively (
As for the 60-chromosome karyotype of Acomys wilsoni, one of the first reported from the Omo Valley by
The spiny mice with 2n=60 were found in central and southern parts of Africa, but the karyotype peculiarities do not allow to consider them conspecific. Comparing to Ethiopian karyotype with FNa=74, two other species with the same 2n=60 show not only differences in FNa – 68 in Acomys selousi De Winton, 1896 and 70 in Acomys spinosissimus – but reveal serious difference in the morphological structure of X chromosome (metacentric with a heterochromatic arm in the first case and submetacentric in the second one). Moreover, they are characterized by the unique XO system in both sexes, accompanied with the atypical inter- and intraindividual autosome number variation (2n=58-62) (
There are still more examples of 2n similarity in the karyotypes of different Acomys. Three of four 2n values that were found in Ethiopia have been reported for Acomys from other territories, i.e. the first finding of 2n=68 from Burkina-Faso (see
The 36-chromosome karyotype is most widely distributed in Mediterranean area and presented among African taxa. The same FN characteristics of the karyotypes with the same 2n may indicate the karyotypic clusters of common origin in the related groups. It is, however, interesting that preliminary data from mt-DNA analysis (Lavrenchenko, in prep.) focused on Ethiopian – i.e. East African – karyotypic forms that we described in this paper do not support their genetic affiliation with taxa from western parts of Africa with the same 2n=40 or 44 and FNa=66 (
Besides the Robertsonian changes occured generally in autosomes, heterochromatin variability exists regarding the X chromosome. To avoid probable variation in FN due to the presence/absence of a short heterochromatic arm in the X-chromosome,
As a result of this preliminary chromosome study, we may conclude that 3 new karyotypes (2n=40, 44, 52) are to be added to at least three karyotypic forms of Ethiopian Acomys (2n=36, 60, 68), described previously (
Karyotypic data on Acomys collected from Ethiopia.
| Species | 2n | FN | FNa | X | Location | Reference |
|---|---|---|---|---|---|---|
| Acomys wilsoni | 60 | 76 | 74 | A | S Ethiopia: Omo Valley |
|
| *Acomys percivali | 36 | 68 | 66 | A | S Ethiopia: Omo Valley |
|
| Acomys cahirinus | 36 | 68 | 66 | A | S Ethiopia: Rift Valley (Konso; Arba-Minch) |
|
| Acomys cahirinus | 36 | 68 | 66 | A | Central Ethiopia: Rift Valley (2 sites along the middle Awash valley) |
|
| Acomys sp. | 68 | 68 | 66 | A | Central Ethiopia: Rift Valley (Koka, upper Awash valley) |
|
| Acomys sp. A | 52 | 68 | 66 | A | NW Ethiopia: Alatish National Park (Amjale) | This study |
| Acomys sp. B | 40 | 68 | 66 | A | NW Ethiopia: Alatish National Park (Bermil) | This study |
| Acomys sp. C | 44 | 68 | 66 | A | E Ethiopia: Babille | This study |
| Total: 6 or more species | 36–68 | 68 (76) | 66 (74) | A | Rift Valley and S, NW and E lowland Ethiopia | 3 publications |
* Referred to Acomys cahirinus in
The authors are indebted to the anonymous referee for valuable criticism of the early version of the submitted manuscript. The supporting grant from the RFBR (Russian Foundation of Basic Researches) is appreciated.