(C) 2011 Marceléia Rubert. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Cytogenetic analyses were performed on fishes of the genus Hypostomus (Hypostomus ancistroides (Ihering, 1911), Hypostomus strigaticeps (Regan, 1908), Hypostomus regani (Ihering, 1905), and Hypostomus paulinus (Ihering, 1905)) from the seven tributaries of the Paranapanema River Basin (Brazil) by means of different staining techniques (C-, Ag-, CMA3- and DAPI-banding) and fluorescence in situ hybridization (FISH) to detect 18S rDNA sites. All species showed different diploid numbers: 2n=68 (10m+26sm+32st-a) in Hypostomus ancistroides, 2n=72 (10m+16sm+46st-a) in Hypostomus strigaticeps, 2n=72 (10m+18sm+44st-a) in Hypostomus regani and 2n=76 (6m+16sm+54st-a) in Hypostomus paulinus. Ag-staining and FISH revealed various numbers and locations of NORs in the group. NORs were usually located terminally on the subtelocentric/acrocentric chromosomes: on the long arm in Hypostomus strigaticeps (2 to 4) and Hypostomus paulinus (2); and on the short arm in Hypostomus ancistroides (2 to 8) and Hypostomus regani (2 to 4). Conspicuous differences in heterochromatin distribution and composition were found among the species, terminally located in some st-a chromosomes in Hypostomus ancistroides, Hypostomus strigaticeps, and Hypostomus paulinus, and interstitially dispersed in most st-a chromosomes, in Hypostomus regani. The fluorochrome staining indicated that different classes of GC and/or AT-rich repetitive DNA evolved in this group. Our results indicate that chromosomal rearrangements and heterochromatin base-pair composition were significant events during the course of differentiation of this group. These features emerge as an excellent cytotaxonomic marker, providing a better understanding of the evolutionary mechanisms underlying the chromosomal diversity in Hypostomus species.
loricariid catfishes, chromosome banding, NORs, fluorochromes, fluorescence in situ hybridization (FISH)
The suckermouth armored catfishes Hypostomus Lacépède, 1803 (Siluriformes, Loricariidae) represent one of the most specious genus of the family Loricariidae, with 127 nominal species (
Most species of this family have a wide distribution in Central and South America. They usually dwell in the rapids, but may be present in different aquatic habitats and in sand banks or rocky rivers. The species of Hypostominae are restricted to freshwater habitats, with the exception of Hypostomus watwata Hancock, 1828, which is a benthic speciesthat lives in estuarine waters. Most of these animals have twilight habits and during daylight hours remain under stones or trunks of dead trees (
The taxonomy of the Loricariidae family has constantly been reviewed through morphological studies (
Among Hypostominae, only eight of its 30 genera (
Concerning the cytotaxonomy, this genus shows a wide variation in diploid number, ranging from 2n=52 in Hypostomus emarginatus Valenciennes, 1840 (
The aim of this work was to analyze specimens of four species of the genus Hypostomus from different populations of the Paranapanema River Basin by means of conventional and molecular cytogenetic techniques and compare the obtained data with the cytogenetic records available for other species of the genus.
A summary of cytogenetic data available for the genus Hypostomus.
| Species | Locality | 2n | FN | KF | NORs | CB | Ref. |
|---|---|---|---|---|---|---|---|
| Hypostomus affinis (Steindachner, 1877) | Jacuí stream (SP) | 66 | 94 | 14m 14sm 12st 26a | 5, t, la | t, la, pc | 9, 10 |
| Hypostomus albopunctatus (Regan, 1908) | Mogi-Guaçu river (SP) | 74 | 104 | 10m 20sm 44st-a | 6, t, sa, la | n.d. | 3 |
| Hypostomus albopunctatus | Piracicaba river (SP) | 74 | 104 | 10m 20sm 44st-a | 3, t, sa, la | i, la, t, sa, pc | 7 |
| Hypostomus ancistroides | n.d. | 68 | 106 | 10m 28sm 30st-a | n.d. | n.d. | 2 |
| Hypostomus ancistroides | Mogi-Guaçu river (SP) | 68 | 102 | 16m 18sm 34st-a | 6, t, sa | n.d. | 3 |
| Hypostomus ancistroides | Araquá river (SP) | 68 | 96 | 18m 10sm 12st 28a | 6, t, sa | n.d. | 12 |
| Hypostomus ancistroides | *** | 68 | 104 | 10m 26sm 32st-a | 6, t, sa | t, la, pc | 16 |
| Hypostomus prope auroguttatus Kner, 1854 | Mogi-Guaçu river (SP) | 76 | 114 | 8m 30sm 38st-a | 2, t, la | n.d. | 3 |
| Hypostomus cochliodon Kner, 1854 | Salobra river and Salobrinha stream (MS) | 64♂ | 100 | 16m 20sm 28st-a | n.d. | t, la | 11 |
| 64♀ | 97 | 16m 19sm 27st-a | n.d. | t, la | 11 | ||
| Hypostomus emarginatus | Araguaia river (MT) | 52 | 98 | 16m 30sm 6st | 2, t, la | n.d. | 5 |
| Hypostomus goyazensis (Regan, 1908) | Vermelho river (GO) | 72 | 98 | 10m 16sm 10st 36a | 2, t, sa | n.d. | 12 |
| Hypostomus macrops (Eigenmann et Eigenmann, 1888) | n.d. | 68 | 92 | 10m 14sm 44st-a | n.d. | n.d. | 2 |
| Hypostomus nigromaculatus (Schubart, 1964) | Mogi-Guaçu river (SP) | 76 | 104 | 8m 20sm 48st-a | 3, t, la | t, la, pc | 15 |
| Hypostomus nigromaculatus | Três Bocas stream (PR) | 76 | 102 | 6m 20sm 50st-a | 3, t, sa, la | t, la, sa, pc | 15 |
| Hypostomus paulinus | n.d. | 74 | 104 | 10m 20sm 44st-a | n.d. | n.d. | 2 |
| Hypostomus paulinus | Três Bocas and Apertados streams (PR) | 76 | 98 | 6m 16sm 54st-a | 2, t, la | t, la, pc | 16 |
| Hypostomus plecostomus (Linnaeus, 1758) | 54 | 90 | 24m 12sm 18st-a | n.d. | n.d. | 1 | |
| Hypostomus regani | Mogi-Guaçu river (SP) | 72 | 102 | 10m 20sm 42st-a | n.d. | n.d. | 3 |
| Hypostomus regani | Araquá river (SP) | 72 | 102 | 12m 18sm 26st 16a | 4, t, la | n.d. | 12 |
| Hypostomus regani | Piumhi river (MG) | 72 | 116 | 8m 16sm 48st-a | 4, t, la | i | 13 |
| Hypostomus regani | Jacutinga river | 72 | 100 | 10m 18sm 44st-a | 4, t, sa | i, pc | 16 |
| Hypostomus strigaticeps | n.d. | 74 | 86 | 8m 4sm 62st-a | n.d. | n.d. | 2 |
| Hypostomus strigaticeps | *** | 72 | 98 | 10m 16sm 46st-a | 4, t, la | t, la, pc | 16 |
| Hypostomus sp. A | Córrego Rincão (SP) | 70 | 102 | 18m 14sm 38st-a | 4, t, sa, la | n.d. | 3 |
| Hypostomus sp. B | Mogi-Guaçu river (SP) | 72 | 102 | 12m 18sm 42st-a | 2, t, la | t, la, pc | 3, 4 |
| Hypostomus sp. C | Mogi-Guaçu river (SP) | 72 | 102 | 10m 18sm 44st-a | 4, t, la | n.d. | 3 |
| Hypostomus sp. D1 | Mogi-Guaçu river (SP) | 72 | 108 | 10m 26sm 36st-a | 4, t, la | n.d. | 3 |
| Hypostomus sp. D2 | Mogi-Guaçu river (SP) | 72 | 106 | 14m 20sm 38st-a | 4, t, la | n.d. | 3 |
| Hypostomus sp. E | Mogi-Guaçu river (SP) | 80 | 104 | 8m 16sm 56st-a | 2, t, sa | t, la, sa, i, pc | 3, 4 |
| Hypostomus sp. F | São Francisco river (MG) | 76 | 102 | 10m 16sm 50st-a | n.d. | pc, t, i | 4 |
| Hypostomus sp. G | Araguaia river (MT) | 64 | 102♂ | 14m 24sm 26st-a | 2, sa | pc, t, i | 6 |
| 64 | 103♀ | 15m 24sm 25st-a | 2, sa | pc, t, i | 6 | ||
| Hypostomus sp.1 | Paranapanema river (SP) | 64 | n.d. | n.d. | n.d. | n.d. | 8 |
| Hypostomus sp.2 | Alambari and Jacutinga streams (SP) | 68 | n.d. | n.d. | n.d. | n.d. | 8 |
| Hypostomus sp. 3 | Quinta and Edgardia stream, Paranapanema river (SP) | 72 | n.d. | n.d. | n.d. | n.d. | 8 |
| Hypostomus sp. 4 | Paranapanema river; Hortelã stream (SP) | 76 | n.d. | n.d. | n.d. | n.d. | 8 |
| Hypostomus sp. 2-rio Perdido NUP 4249 | Perdido river (MS) | 84 | 106 | 6m 16sm 62st-a | 2, t, la | pc, t, la | 14 |
| Hypostomus sp. 3-córrego Salobrinha NUP 4247 | Salobra river and Salobrinha stream (MS) | 82 | 102 | 6m 14sm 62st-a | 2, t, la | pc, t, la | 14 |
| Hypostomus sp.1a | Patos stream (MG) | 76 | 106 | 6m 8sm 62st-a | 3, t, sa, la | t, la | 13 |
| Hypostomus sp.1b | Araras stream (MG) | 76 | 106 | 6m 8sm 62st-a | 3, sa, la | t, la | 13 |
| Hypostomus sp.2 | Araras stream (MG) | 74 | 106 | 10m 6sm 58st-a | 2, la | t, la | 13 |
Cytogenetic analysis was performed on a total of 148 specimens of four Hypostomus species collected at different sites of the Paranapanema River Basin (southern Brazil) (Table 1). The specimens were deposited in the Museu de Zoologia of the Universidade Estadual de Londrina (MZUEL), Londrina, Paraná State, Brazil.
Conventional stainingMetaphase chromosomes were obtained through the air-drying technique (
C-banding was performed according to
The fluorescence in situ hybridization procedure was performed according to
Images were acquired with Leica DM 4500 B microscope equipped with a DFC 300FX camera and Leica IM50 4.0 software.
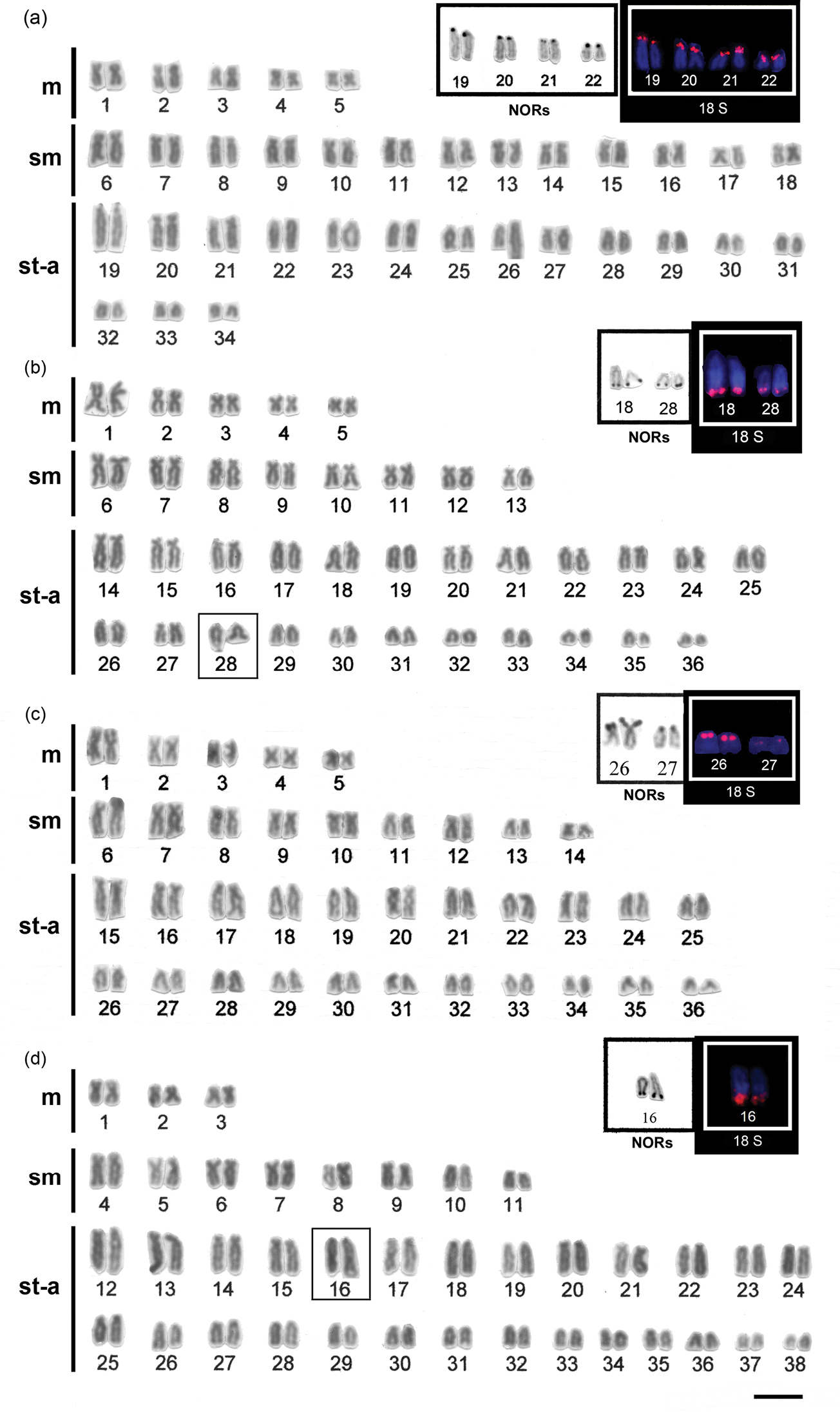
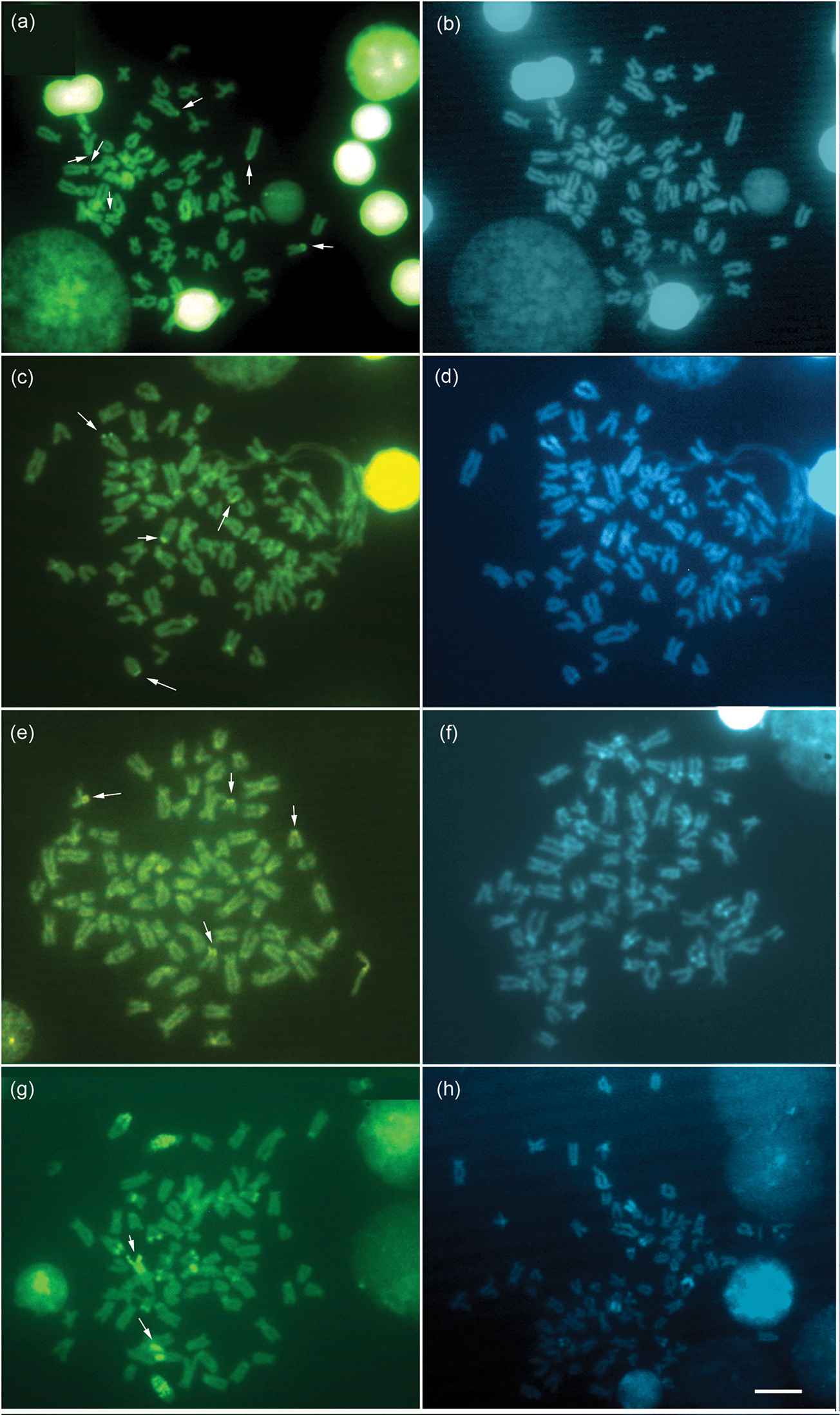
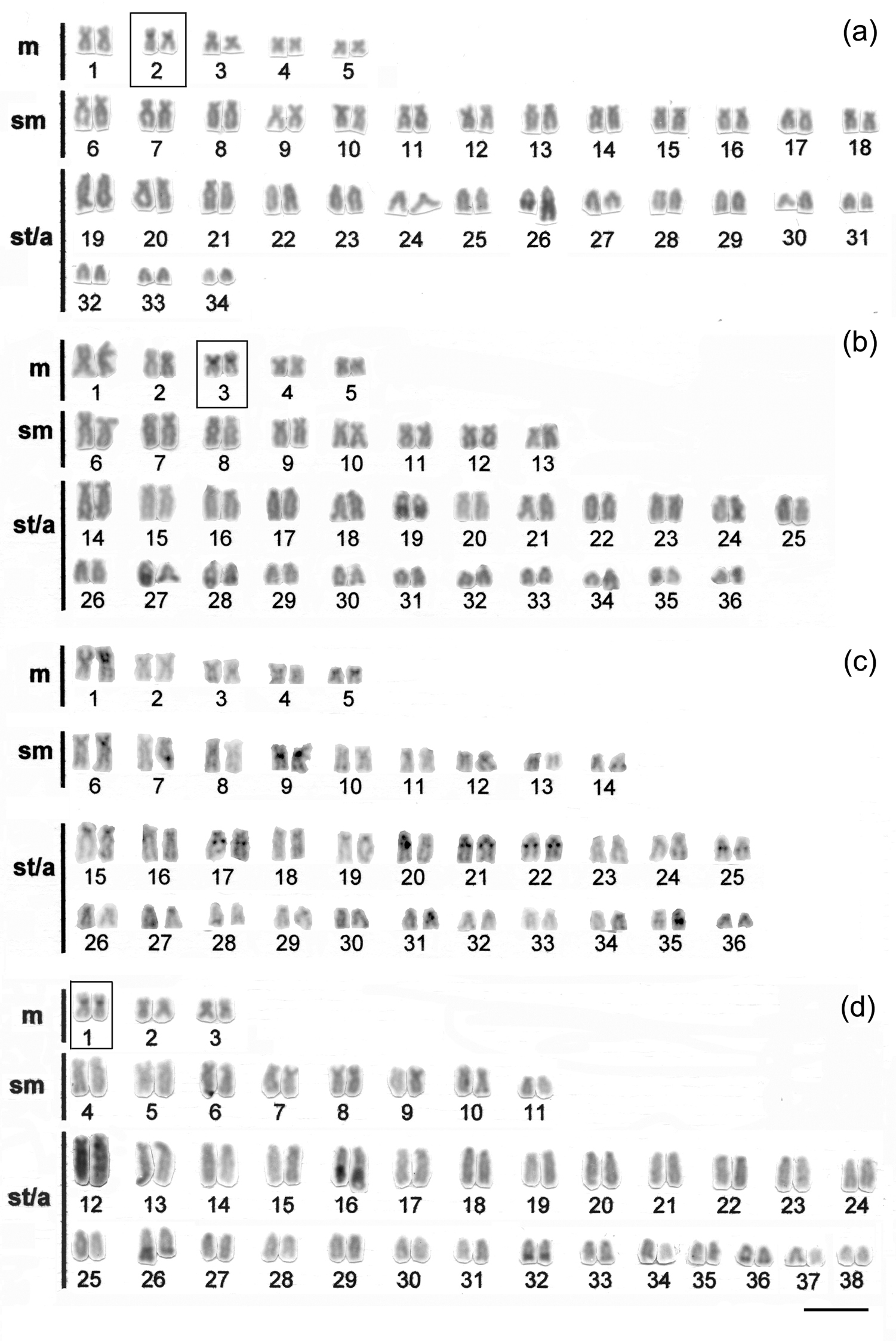
ResultsSpecimens of Hypostomus ancistroides showed a diploid number 2n=68 and a fundamental number (FN) of 104, with a karyotype formula of 10m+26sm+32st-a. One chromosome of pair 26 showed size heteromorphism (Fig. 1a). Silver nitrate staining (Fig. 1a left box) and FISH (Fig. 1a right box) revealed up to four pairs of subtelocentric/acrocentric NOR-bearing chromosomes. CMA3 marked the terminal region of the long arms of pair 26, the pericentromeric region of the second pair of metacentric chromosomes, and probably the NOR-bearing chromosomes (Fig. 2a). No fluorescent staining was observed after DAPI staining (Fig. 2b). Heterochromatin was distributed in the pericentromeric region of the second pair (m) of the complement and in the terminal region of the long arm (pair 26) (Fig. 3a).
Hypostomus strigaticeps presented a diploid number 2n=72 and a FN of 98, with a karyotype formula of 10m+16sm+46st-a (Fig. 1b). The Ag-NOR site numbers ranged from two to four marked chromosomes (st-a) located in the terminal region of the long arm (pairs 18 and 28) (Fig. 1b left box), similar to the number observed in FISH (Fig. 1b right box). CMA3 marked four chromosomes, possibly the Ag-NOR sites, and the pericentromeric regions of most subtelocentric/acrocentric chromosomes (Fig. 2c). Staining with DAPI revealed large blocks in the terminal regions of four-eight subtelocentric/acrocentric chromosomes (Fig. 2d). C-banding revealed the occurrence of heterochromatic blocks in the pericentromeric region of the third pair of metacentric chromosomes and of up to eight large blocks in the terminal regions of the long arms of subtelocentric/acrocentric chromosomes. In one of those chromosome pairs, the heterochromatic block was adjacent to the secondary constriction (Fig. 3b).
Hypostomus regani had 2n=72 with a karyotype formula of 10m+18sm+44st-a and FN of 100 (Fig. 1c). Ag-NORs were located in the terminal position on the short arms of four subtelocentric/acrocentric chromosomes (pairs 26 and 27) (Fig. 1c left box). The same number of NOR-bearing chromosomes was observed after FISH (Fig. 1c right box) and CMA3-staining (Fig. 2e). Interstitial CMA3-negative blocks were observed in most of the subtelocentric/acrocentric chromosomes, which, in contrast, were positive after DAPI staining (Fig. 2f). Heterochromatin was distributed in the interstitial region of most st-a chromosomes and in the pericentromeric region of one metacentric pair (Fig. 3c).
Hypostomus paulinus showed 2n=76, FN=98 and a karyotype formula of 6m+16sm+54st-a (Fig. 1d). NORs were located in the terminal position on the long arms of chromosome pair 16 (Fig. 1d left box), similar to the chromosomes observed in FISH (Fig. 1d right box). CMA3-banding marked up to eight chromosomes (st-a) with large GC-rich blocks, and one st-a pair, probably corresponding to NOR-bearing chromosomes, and in the pericentromeric region of the first (m) pair (Fig. 2g); after DAPI staining, eight fluorescent bands were observed (Fig. 2h). Heterochromatin was distributed in the pericentromeric region of the first pair of metacentric chromosomes, in the terminal region of the long arms of eight pairs of subtelocentric/acrocentric chromosomes, one of which was the NOR-bearing pair. In this pair, a heterochromatin block was located at the proximal portion of the secondary constriction, whereas three heterochromatin blocks, which occupied almost the entire long arm, were observed in a pair of subtelocentric/acrocentric chromosomes (pair 12) (Fig. 3d).
Karyotypes of a Hypostomus ancistroides b Hypostomus strigaticeps c Hypostomus regani d Hypostomus paulinus arranged from Giemsa-stained chromosomes. In the insets, partial karyotypes of the NOR-bearing chromosome pairs after Ag-staining (left) and FISH with 18S rDNA probe (right). Bar = 10 µm.
Metaphases stained with CMA3 (left) and DAPI (right), of Hypostomus ancistroides a, b Hypostomus strigaticeps c, d Hypostomus regani e, f Hypostomus paulinus g, h. The arrows indicate the NOR-bearing chromosomes. Bar = 10 µm.
Karyotypes of a Hypostomus ancistroides b Hypostomus strigaticeps c Hypostomus regani and d Hypostomus paulinus, arranged from C- banded chromosomes Bar = 10 µm.
All species differed with respect to their diploid chromosome number and/or karyotype, as follows: 2n=68 (10m+26sm+32st-a) in Hypostomus ancistroides (Fig. 1a), 2n=72 (10m+16sm+46st-a) in Hypostomus strigaticeps (Fig. 1b), 2n=72 (10m+18sm+44st-a) in Hypostomus regani (Fig. 1c), and 2n=76 (6m+16sm+54st-a) in Hypostomus paulinus (Fig. 1d). This variability is consistent with the chromosomal data previously reported in the genus Hypostomus, which showed a wide variation in 2n (from 52 to 84) (Table 1). The available cytogenetic studies showed that the species that possess the same 2n have different karyotypes. In the same way as the features observed in Hypostomus ancistroides (2n=68) but with different fundamental numbers (FN) among different populations, i.e. 106, 102 and 96 (
According to
The same variability found in 2n and in karyotypes was also detected in NORs. Our data showed different phenotypes among the Hypostomus species, observed after silver staining and FISH. All species showed Ag-NORs and 18S rDNA sites located in the terminal regions of st-a chromosomes, but with a significant variation in number and location among them. Hypostomus ancistroides showed up to 8 NOR sites, all located on the short arms (Fig. 1a left and right boxes, respectively). Hypostomus strigaticeps showed NORs on the long arms and Hypostomus regani, NORs located on the short arms, and both species with up to 4 sites (Fig. 1b and 1c left and right boxes, respectively), and Hypostomus paulinus evidenced only two NOR-bearing chromosomes located on the long arm (Fig. 1d left and right boxes), which could be considered as species-specific characteristics.
The presence of one pair of NOR-bearing chromosomes, and also its interstitial location seems to be a widespread condition for Loricariidae fish, since this occurs among the Neoplecostominae and Hypoptopomatinae species (
In the four species presently studied, the NORs were positive for CMA3 staining (Fig. 2), a feature that has been conserved among all Neoteleostei (
Some other studies carried out in Hypostomus (
The chromosome banding performed in all species analyzed showed a variation in the heterochromatin distribution pattern. However, the presence of heterochromatin in some chromosomes was constant, as observed in the pericentromeric region of a metacentric pair in Hypostomus ancistroides (pair 2), Hypostomus strigaticeps (pair 3), and Hypostomus paulinus (pair 1) (Fig. 3a, b, d, respectively), also reported in Hypostomus nigromaculatus by
The presence of a marker chromosome that seems conserved for most Hypostomus species, corresponding to the NOR-bearing chromosome pair, which shows a heterochromatin block adjacent to this site (e.g.
Karyotypes, banding patterns, number and location of ribosomal DNA sites, and repetitive DNA are important tools for the cytotaxonomy of Hypostomus species. Since these characteristics do not vary among the different populations of the same species, they are significant cytogenetic markers at the species level.
Further data on other Hypostomus species from different rivers, as well as detailed studies of satellite DNA sequences may clarify important issues of genome organization, be used as genetic markers, and provide interesting insights for the comprehension of the evolution of this genus.
The authors are grateful to Dr. Ana Lúcia Dias and Dr. André L. L. Vanzela for the review of this manuscript. This research was supported by a grant from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).