(C) 2012 Marcela Maria Pereira de Lemos Pinto. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
The genus Artibeus represents a highly diverse group of bats from the Neotropical region, with four large species occurring in Brazil. In this paper, a comparative cytogenetic study was carried out on the species Artibeus obscurus Schinz, 1821, Artibeus fimbriatus Gray, 1838, Artibeus lituratus Olfers, 1818 and Artibeus planirostris Spix, 1823 that live sympatrically in the northeast of Brazil, through C-banding, silver staining and DNA-specific fluorochromes (CMA3 and DAPI). All the species had karyotypes with 2n=30, XX and 2n=31, XY1Y2, and FN=56. C-banding showed constitutive heterochromatin (CH) blocks in the pericentromeric regions of all the chromosomes and small CH blocks at the terminal region of pairs 5, 6, and 7 for all species. Notably, our C-banding data revealed species-specific autosomic CH blocks for each taxon, as well as different heterochromatic constitution of Y2 chromosomes of Artibeus planirostris. Ag-NORs were observed in the short arms of chromosomes 5, 6 and 7 in all species. The sequential staining AgNO3/CMA3/DA/DAPI indicated a positive association of CH with Ag-NORs and positive CMA3 signals, thus reflecting GC-richness in these regions in Artibeus obscurus and Artibeus fimbriatus. In this work it was possible to identify interespecific divergences in the Brazilian large Artibeus species using C-banding it was possible provided a suitable tool in the cytotaxonomic differentiation of this genus.
C-banding, Ag-NOR, CMA3/DA/DAPI , Cytotaxonomy
The genus Artibeus Leach, 1821 has been divided into two main groups based on body size. The species with larger body size have been classified as subgenus Artibeus and the species with smaller body size as the subgenus Dermanura Gervais, 1856. In addition, a new subgenus Koopmania Owen, 1991 was proposed by
The extensive similarity of morphometric characters, high degree of shape diversity and overlapping of natural habitats have hindered accurate identification of the large Artibeus along their distribution, particularly in the Neotropical region (
Since the systematic classification of subgenus Artibeus remains subject of several discussions concerning phylogenetic relationships and actual taxonomic status of species, the use of complementary information may help to define species more precisely (
In this work a karyotypic characterization of Artibeus obscurus Schinz, 1821, Artibeus fimbriatus Gray, 1838, Artibeus lituratus Olfers, 1818 and Artibeus planirostris Spix, 1823 from northeastern of Brazil was performed by the cytogenetic techniques – conventional analysis, C-banding, Ag-NOR and triple staining CMA3/DA/DAPI. The data were helpful to carry a comparative analysis of those species, in terms of interspecific differences, and also to provide a better identification of them.
Material and methodsBased on literature (
After identification, cytogenetic studies were carried out on 53 Artibeus specimens from the state of Pernambuco, northeastern Brazil. Voucher specimens are deposited in the Mammalian collection at the Department of Systematic and Ecology, Federal University of Paraíba, João Pessoa, Paraiba, Brazil. The specimens studied were six males and eight females of Artibeus obscurus; two males and four females of Artibeus fimbriatus; eight males and five females of Artibeus planirostris; ten males and ten females of Artibeus lituratus captured at different sites across the Pernambuco State: Igarassu (07°50'02"S, 34°54'21"W), Água Preta (08°42'27"S, 35°31'50"W), Rio Formoso (08°39'50"S, 35°09'32"W), Ipojuca (08°24'00"S, 35°03'45"W) and Recife (08°03'14"S, 34°52'51"W) (see also Appendix).
Metaphase spreads were obtained from bone marrow cells according to conventional procedures and staining with Giemsa. C-banding and silver staining were performed according to
For sequential staining (AgNO3/CMA3/DA/DAPI), the slides stained by silver nitrate were distained after photographing (
All four species shared the same diploid number (2n=30, gap XX; 2n=31, gap XY1Y2) and fundamental number FN=56. Chromosomes were meta-submetacentric (1-4, 8-14), subtelocentric (5, 6, 7 and X) and two small acrocentric (Y1 and Y2). Except for the size of Y1 and Y2 chromosomes, it was not found any intraspecific variation between species analyzed with conventional staining.
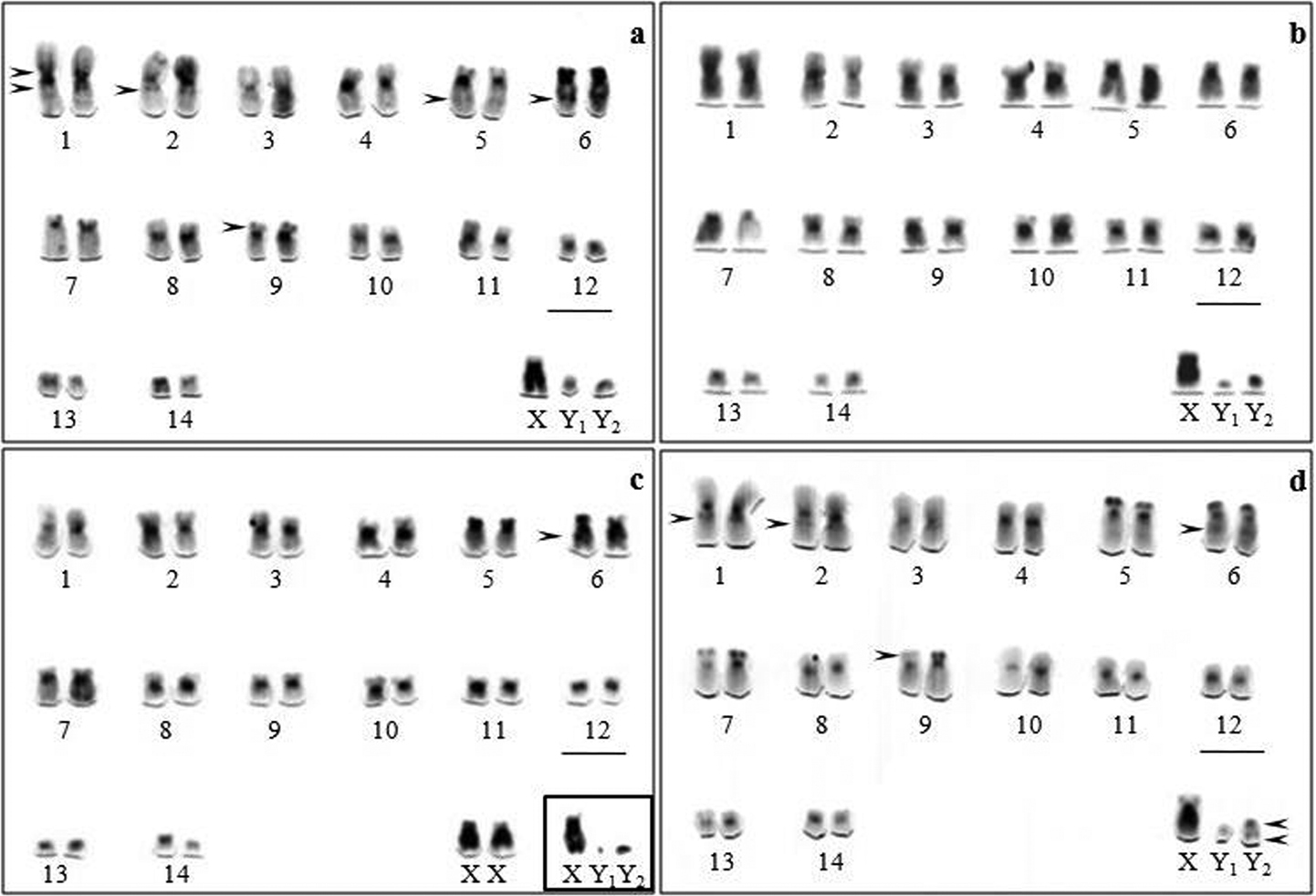
C-banding revealed constitutive heterochromatin (CH) in the pericentromeric region of all the autosomes and small heterochromatic blocks were observed in the terminal region of chromosome pairs 5, 6 and 7 (Fig. 1a–d). The karyotype of Artibeus obscurus (Fig. 1a) exhibited interstitial blocks in the short and long arms of pair 1, as well as in the long arms of pairs 2, 5, 6 and in the terminal region of the short arm of pair 9. The Artibeus planirostris karyotype (Fig. 1d) has the same CH pattern but lacks interstitial blocks in the short arm of chromosome 1. Absence of an interstitial block on the chromosome 6 distinguished karyotype of Artibeus fimbriatus from the other investigated karyotypes (Fig. 1). In all the material examined the long arms of the X chromosomes were more darkly stained when compared with the euchromatin of the autosomes. The Y2 appeared almost entirely heterochromatic in all species, except for Artibeus planirostris which showed pericentromeric and distal blocks (Fig. 1d). The pattern of the Y1 could not be determined with precision due to its punctiform size.
C-banding of Artibeus obscurus (a) Artibeus fimbriatus (b) Artibeus lituratus (c) and Artibeus planirostris (d) karyotypes. The arrowheads indicate a particular set of CH blocks in each species. Bar = 5 µm.
Table 1 shows exhibits the C-banding pattern in chromosomal complement in all species analyzed.
Heterochromatin pattern in chromosomal complement in Artibeus species
| Species | C-banding | ||||
| Pericentromeric | Terminal | Interstitial | Dispersed | Distal | |
| Artibeus obscurus | + | 5p, 6p, 7p, 9p | 1*, 2q, 5q, 6q | Y1 e Y2 | - |
| Artibeus fimbriatus | + | 5p, 6p, 7p | - | Y1 e Y2 | - |
| Artibeus lituratus | + | 5p, 6p, 7p | 6q | Y1 e Y2 | - |
| Artibeus planirostris | + | 5p, 6p, 7p, 9p | 1q, 2q, 5q, 6q | Y1 | Y2 |
Silver staining (Ag-NORs) showed three pairs of NORs in the terminal region of the short arms in chromosomes 5, 6 and 7 in all species. As a result of remarkable variation in expression and activity, Ag-NORs were counted up to 100 nuclei, which were randomly selected, and the mean number of Ag-NORs per nucleus was determined for each case (Table 2).
Frequency analyzes of active NORs in the large species of genus Artibeus.
| Species | Active NOR number per cell | Total of cells analyzed | |||||
| 1 | 2 | 3 | 4 | 5 | 6 | ||
| Artibeus obscurus | 0 | 17 | 53 | 71 | 12 | 16 | 169 |
| Artibeus fimbriatus | 0 | 14 | 40 | 58 | 27 | 22 | 161 |
| Artibeus lituratus | 0 | 7 | 30 | 36 | 22 | 24 | 119 |
| Artibeus planirostris | 0 | 14 | 28 | 47 | 11 | 25 | 125 |
| Total | 0 | 52 | 151 | 212 | 72 | 87 | 574 |
| (%) total | 0 | 9.06 | 26.31 | 36.94 | 12.54 | 15.16 | |
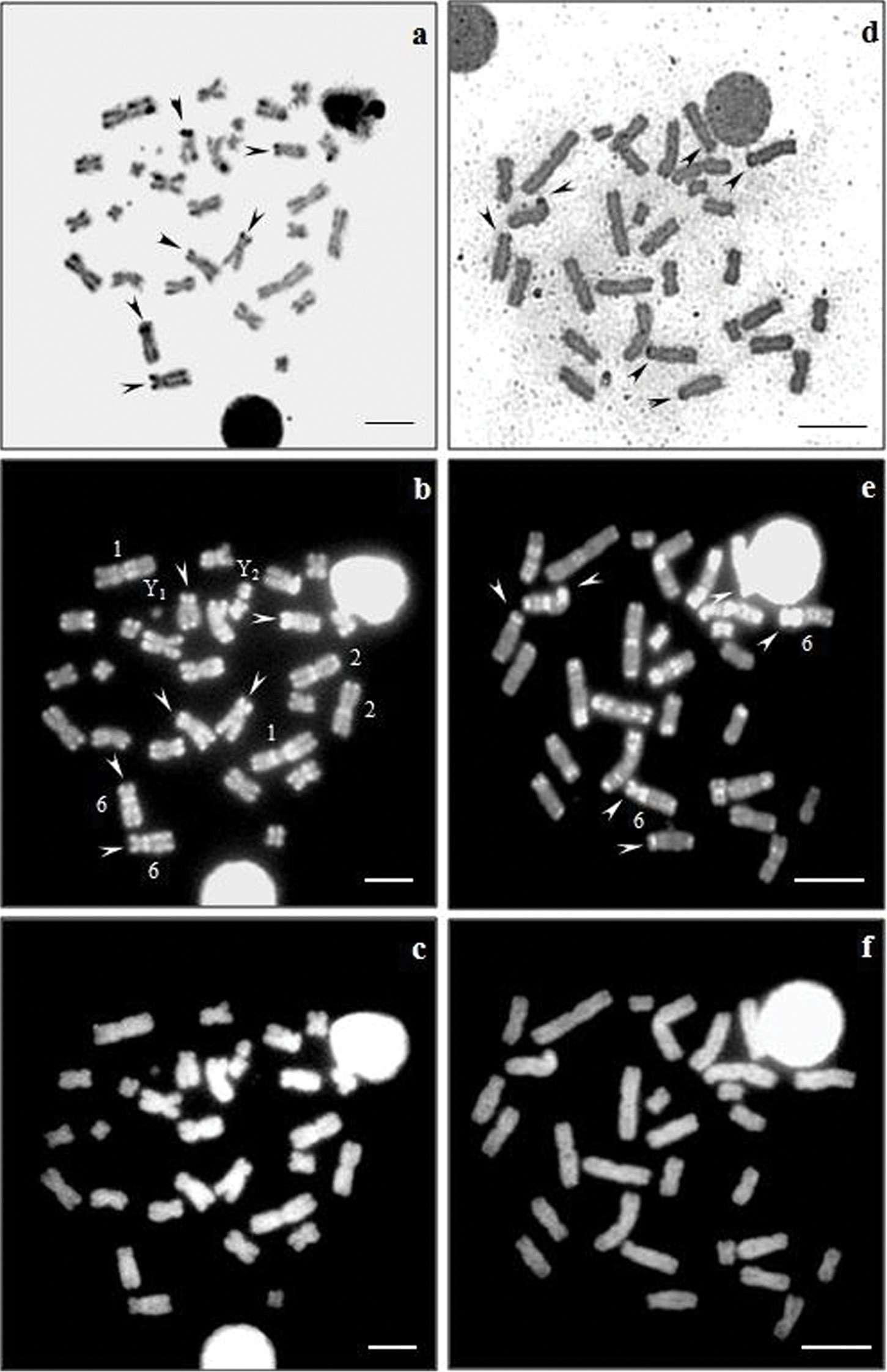
The sequential staining AgNO3/CMA3/DA/DAPI showed a correlation between CMA3 positive regions and Ag-NORs in the karyotypes of Artibeus obscurus and Artibeus fimbriatus (Fig. 2a–f). Karyotypes of both species had presented CH blocks associated with Ag-NORs sites, reflecting GC-richness in these heterochromatics clusters. In addition, positive CMA3 signals were observed in the pericentromerics regions of certain autosomes, particularly in pairs 1, 2 and 6 of Artibeus obscurus and in pair 6 of Artibeus fimbriatus (Fig. 2b, e). On the other hand, a uniform pattern was observed in all the chromosomes after DA/DAPI staining (Fig. 2c–d).
Sequential staining of Artibeusobscurus (a–c) and Artibeus fimbriatus (d–f) karyotypes with AgNO3/CMA3/DA/DAPI. (a, d) Ag-NORs, (b, e) CMA3, (c–f) DA/DAPI. Bar = 5 µm.
Our data regarding diploid number, chromosome morphology and sex determination system obtained for Artibeus obscurus, Artibeus fimbriatus, Artibeus lituratus and Artibeus planirostris karyotypes are in agreement with those previously described in the literature (
The multiple sex chromosome system XY1Y2 has been widely reported within the genus Artibeus, e.g. Artibeus aztecus Andersen, 1906, Artibeus glaucus Thomas 1893, Artibeus toltecus, Artibeus concolor, Artibeus cinereus Gervais, 1856, Artibeus hirsutus Andersen, 1906, Artibeus inopinatus Davis et Carter, 1964 and Artibeus jamaicensis, and for other 23 species of family Phyllostomidae, as predominant type of sex-determining mechanism in this group (
The CH distribution was evaluated and intercompared in the large Artibeus and with others phyllostomatids, pointing out an extensive similarity of CH pattern localized in the pericentromeric region (
On the other hand, a particular set of CH blocks was observed in Artibeus obscurus, Artibeus fimbriatus, Artibeus lituratus and Artibeus planirostris. This finding allowed the individualization and differentiation of each species for karyotype comparison (Fig. 1). Artibeus fimbriatus and Artibeus lituratus karyotypes showed a closer CH distribution differing only by one heterochromatin block. Furthermore Artibeus obscurus and Artibeus planirostris karyotypes presented more interstitial heterochromatin.
The occurrence of intrageneric variation on CH distribution had been described only in sporadic cases among phyllostomatids whose extensive karyotypic conservation is widely known. In turn, the genus Artibeus is widely cited as a chiropteran group that exhibits low rate of karyotype evolution whereas: (1) most of species had same diploid number (30/31) and (2) G-banding patterns are essentially identical (
The other parameter evaluated intercomparison was the NORs localization by silver staining. The Ag-NORs were situated on the subtelocentric autosomes 5, 6 and 7 of all species. The data obtained for Artibeus lituratus, Artibeus planirostris and Artibeus fimbriatus, together with the new data of Artibeus obscurus, were similar those described by
As only active NORs could be visualized in our data, the variation in Ag-NORs activity for cell was also investigated (Table 2). In the most of cells analyzed (> 500), the frequency of active NORs was 3 or 4 black spots (26.31 to 36.94 %). Such variability is in accordance with other studies on a NOR sites activity in Phyllostomidae bats that presents multiple NORs (
The association between NORs and CH by GC-specific fluorochromes staining presented in this work for Artibeus obscurus and Artibeus fimbriatus, has also been reported to Artibeus lituratus, Artibeus jamaicencis, Desmodus rotundus Geoffroy, 1810, Diphylla ecaudata Spix, 1823 and Lonchorhina aurita Tomes, 1863. On the other hand, Carollia perspicillata Linnaeus, 1758, Molossus molossus Pallas, 1766, Molossus ater Peters, 1865, Molossops planirostris Peters, 1865, Phyllostomus discolor Wagner, 1843 and Trachops cirrhosus Spix, 1823 NORs and CH were CMA3 neutral. The reason for that is probably in heterogeneity of base composition of the intergenic regions related to NORs. In some cases, the triple staining with CMA3/DA/DAPI has also enhanced the patterns of R-bands with CMA3, an uniform staining with DA/DAPI or a weak G-banding pattern, as it has been observed in some bat’s families (
Classical and molecular cytogenetic markers, associated to taxonomic studies, have provided a better understanding of phylogenetic relationships and the mechanisms responsible for chromosomal divergence in the different taxa in the order Chiroptera. Cytogenetic analysis of all Brazilian species of the subgenus Artibeus allowed us to reveal the conservative and specific chromosomal features among their karyotypes. Furthermore, it was possible to identify intrageneric and interespecific divergences in a group that up to today has been characterized by showing extensive karyotypic conservation. The cytogenetic techniques herein employed, demonstrated the usefulness of C-banding in the identification and correct individualization of the large Artibeus that live sympatrically in the northeastern of Brazil, thus providing an important tool in the cytotaxonomic differentiation of this genus.
The authors are grateful to Francisca Tavares Lira and Cirlene Maria da Silva to provide technical support and to Ana Emília Barros e Silva to helping in the use of the System image capture. We are thankful to Cibele Gomes de Sotero-Caio for the final review of the manuscript. This Project had financial support of the CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico).
Família Phyllostomidae
Artibeus obscurus
M 318 (3214) – Saltinho (Rio formoso)
M 319 (3206) – Saltinho (Rio formoso)
M 336 (3235) – Saltinho (Rio formoso)
M 337 (3238) – Saltinho (Rio formoso)
M 340 (3216) – Saltinho (Rio formoso)
M 344 (3237) – Saltinho (Rio formoso)
M 359 (3230) – Saltinho (Rio formoso)
M 381 (3181) – Saltinho (Rio formoso)
M 397 (3186) – Dois irmãos (Recife)
M 455 (3189) – Saltinho (Rio formoso)
M 478 (3179) – Saltinho (Rio formoso)
Artibeus fimbriatus
M 346 (3215) – Saltinho (Rio formoso)
M 382 (3184) – Saltinho (Rio formoso)
M 395 (3177) – Dois irmãos (Recife)
M 453 (3175) – Saltinho (Rio formoso)
M 479 (3192) – Saltinho (Rio formoso)
Artibeus lituratus
M 221 (3418) – Igarassu
M 379 (3178) – Saltinho (Rio formoso)
M 446 (3191) – Saltinho (Rio formoso)
M 454 (3182) – Saltinho (Rio formoso)
M 475 (3188) – Saltinho (Rio formoso)
M 476 (3180) – Saltinho (Rio formoso)
Artibeus planirotris
M 118 (3424) – Igarassu
M 124 (3212) – Igarassu
M 137 (3423) – Aldeia (Camaragibe)
M 188 (3428) – Igarassu
M 262 (3220) – Água Preta (Fazenda Camarão)
M 263 (3218) – Água Preta (Fazenda Camarão)
M 393 (3202) – Dois irmãos (Recife)
M 400 (3176) – Dois irmãos (Recife)
M 401 (3196) – Dois irmãos (Recife)