(C) 2012 Valéria Pereira Mendes. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Cytogenetic studies were performed on the species Apteronotus prope albifrons Linnaeus, 1766, Rhamphichthys hahni Meinken, 1937 and Brachyhypopomus gauderio Giora & Malabarba, 2009, collected in the upper Paraná River floodplain, Porto Rico (PR), Brazil. Apteronotus prope albifrons showed a diploid number of 2n=24 chromosomes for both sexes and a karyotype formula of 14m+2sm+2st+6a (FN=42). Besides the standard karyotype, three specimens had one to three extra microchromosomes with inter- and intra-individual variations, which suggested the occurrence of B chromosomes in the species. The chromosomal data of Rhamphichthys hahni, described here for the first time, consists of 50 chromosomes and a formula comprised of 20m+24sm+6a (FN=94). Brachyhypopomus gauderio specimens demonstrated 2n=42 chromosomes infemales, all acrocentric, and 2n=41 chromosomes in males, with 40 acrocentric and 1 medium-sized metacentric chromosome. These differences concern with a multiple system of sexchromosome determination X1X1X2X2/X1X2Y (FN=42) in Brachyhypopomus gauderio. The analysis of nucleolar organizer regions by Ag-NOR and FISH 18S banding revealed a simple NOR system in Apteronotus prope albifrons and Rhamphichthys hahni and a multiple NOR system in Brachyhypopomus gauderio, that is unusual for Gymnotiformes fishes. Constitutive heterochromatin was mainly found in the pericentromere region in most of the chromosomes of the three species, although each species had its own peculiarities. The B chromosomes in Apteronotus prope albifrons demonstrated heterochromatin positioned in the centromeric and telomeric regions whereas Rhamphichthys hahni presented conspicuous blocks of heterochromatin on the long arms in three submetacentric pairs. Brachyhypopomus gauderio showed blocks of heterochromatin on the long arm in the interstitial and telomere positions. The finding of B chromosomes in Apteronotus prope albifrons represents the first description of these elements in the Gymnotiformes order. Although the karyotype of this species is similar with that described for populations in the Amazon basin, the presence of B chromosomes could represent a specific characteristic of this population. A comparative analysis of karyotypes of Rhamphichthys hahni with other species of the genus showed a relatively conservative structure suggesting 2n=50 as a common number in this group. The karyotype of Brachyhypopomus gauderio, a new species, provides an important reference for future chromosome studies of the Brachyhypopmus Mago-Lecia, 1994, and it might be also significant for cytotaxonomy in this group. The cytogenetic data also demonstrate the need of more comparative cytogenetic studies in the families of the highly diversified and taxonomically difficult complex Gymnotiformes.
fish cytogenetics, B chromosomes, C-banding, ribosomal DNA, sex chromosomes
The order Gymnotiformes is a group with high species diversity and about 179 species have been listed so far (
Karyotype diversity is well known in Gymnotiformes, especially in the genera Gymnotus Linnaeus, 1758 and Eigenmannia Jordan et Evermann, 1896. Regarding this order, the diploid number ranges from 2n = 22 or 24 in Apteronotus albifrons Linnaeus, 1766 (
The karyotype variability in Gymnotiformes also involves simple and multiple sex chromosome systems such as those registered in Gymnotidae, Hypopomidae and Sternopygidae species. The sex chromosome systems XX/XY and ZZ/ZW were described in Eigenmannia virescens Valenciennes, 1836 (
Cytogenetic data available for Apteronotidae, Rhamphichthyidae and Hypopomidae families. FN: fundamental number; m: metacentric; sm: submetacentric; st: subtelocentric; a: acrocentric; AM: Amazonas; PR: Paraná; SP: São Paulo; PA:Pará.
| Family/species | Locality | 2n | FN | Karyotype | Sex chromo-somes | Reference |
|---|---|---|---|---|---|---|
| Apteronotidae | ||||||
| Apteronotus albifrons | Amazon | 24 | 42 | 14m+2sm+2st+6a |
|
|
| Apteronotus albifrons | Marajó Island, PA | 24 | 42 | 12m+4sm+2st+6a |
|
|
| Apteronotus prope albifrons | Upper PR, PR | 24 | 42 | 14m+2sm+2st+6a | Present study | |
| Apteronotus albifrons | 22 |
|
||||
| Apteronotus sp. | São Paulo | 52 | 98 | 46m/sm+ 6st/a |
|
|
| Parapteronotus bonaparti (Apteronotus anas) | Manaus (AM) | 52 | 94 | 30m+12sm+10a |
|
|
| Parapteronotus hasemani (Apteronotus) | Manaus, AM | 52 | 94 | 26m+16sm+10a |
|
|
| Rhamphichthyidae | ||||||
| Rhamphichthys hani | Upper PR, PR | 50 | 94 | 20m+24sm+6a | Present study | |
| Rhamphichthys prope pantherinus | Amazon | 52 | 100 | 38m+10sm+4st |
|
|
| Rhamphichthys marmoratus | Amazon | 50 | 94 | 44m/sm+6st/a |
|
|
| Rhamphichthys rostratus | Amazon | 50 | 92 | 42m/sm+8a |
|
|
| Hypopomidae | ||||||
| Brachyhypopomus brevirostris | Amazon | 36 | 42 | 4m+2sm+8st+22a |
|
|
| Brachyhypopomus gauderio | Upper PR, PR | ♀ 42 | 42 | 42a | X1X1X2X2 | Present study |
| ♂ 41 | 42 | 1m+40a | X1X2Y | |||
| Tietê River, SP | ♀ 42 | 42 | 42a | X1X1X2X2 |
|
|
| ♂ 41 | 42 | 1m+40a | X1X2Y | (as Brachyhypopomus pinnicaudatus) | ||
| Hypopomus artedi | Amazon | 38 | 70 | 32m/sm+6st/a |
|
|
| Hypopygus lepturus | Amazon | 50 | 96 | 16m+20sm+10st+4a |
|
|
In spite of extensive karyotype variability, a review of the cytogenetic studies of Neotropical freshwater fish shows that, to date, there is no record of B chromosomes (or supernumerary chromosomes) in Gymnotiformes (
Although Neotropical fishes show considerable variability regarding the number of B chromosomes (1–16), usually 1–4 chromosomes are present. The B chromosomes show wide variations in size, from very small (micro), as in Moenkhausia sanctaefilomenae Steindachner, 1907 (
Methodologies such as C-banding have revealed the nature of heterochromatic B chromosomes with repetitive DNA sequences (
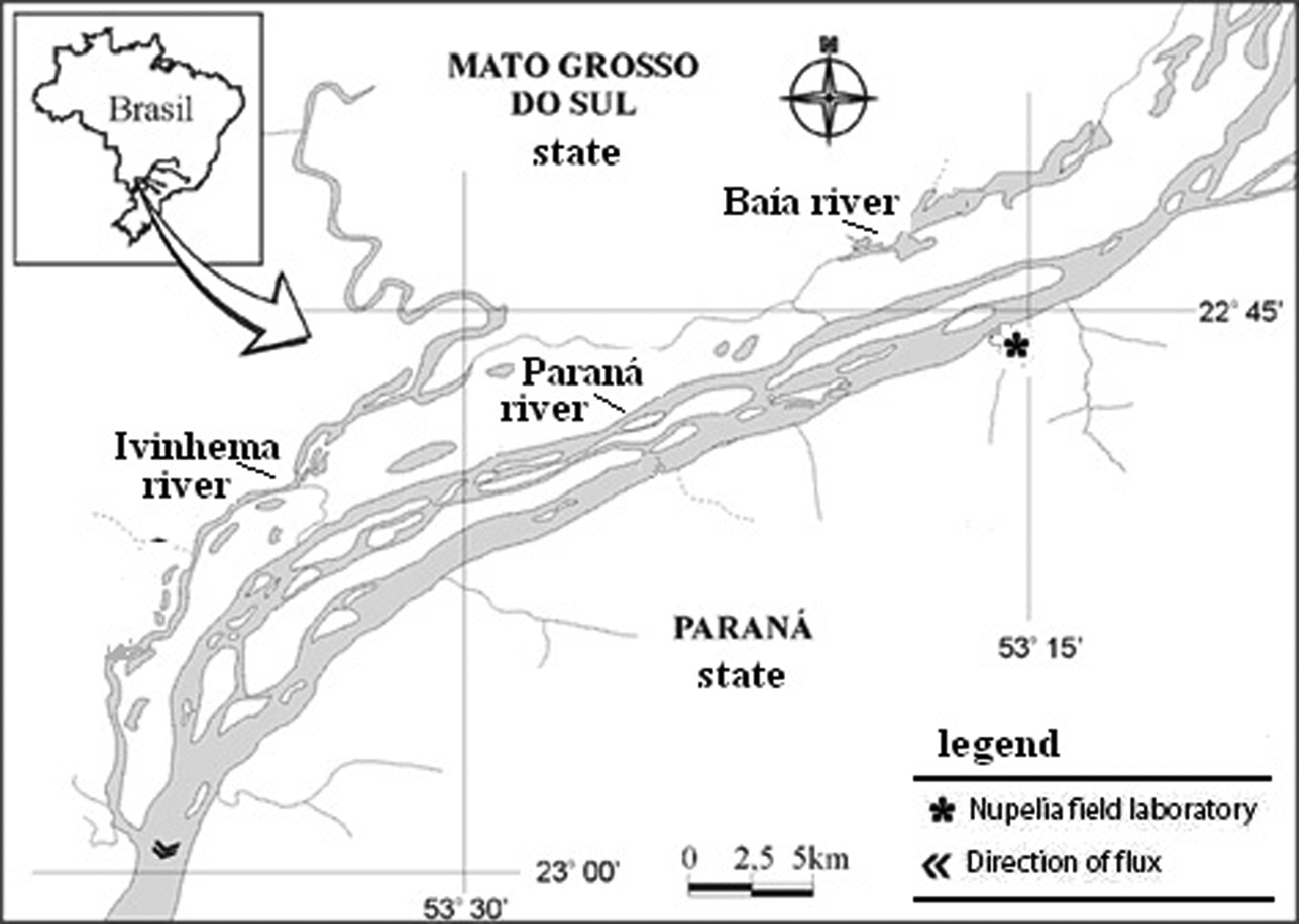
We analysed 51 specimens (24 males and 27 females) of Brachyhypopomus gauderio, 6 specimens of Apteronotus prope albifrons (4 males and 2 females) and 19 specimens of Rhamphichthys hahnii (8 males, 7 females and 4 undetermined sex). The specimens were collected in rivers (Baía and Ivinhema) and lagoons of the upper Paraná River floodplain near the town of Porto Rico (PR), Brazil (Fig. 1). Voucher specimens were deposited in the fish collection of the Research Nucleus in Limnology, Ichthyology and Aquaculture (Nupélia), Universidade Estadual de Maringá, PR Brazil, as Apteronotus prope albifrons (NUP9621), Rhamphichthys hahni (NUP9623) and Brachyhypopomus gauderio, (earlier misidentified as Brachyhypopomus prope pinnicaudatus, NUP9622). Brachyhypopomus gauderio represents a new species of the southern Brazil, Uruguay and Paraguay described by
Collections sites of studied species. Map of Brazil showing the localization of Paraná river, in the Paraná state. Hhydrographic map of the floodplain of the Upper Paraná River and its tributaries, Baía and Ivinhema Rivers.
Specimens of: a Apteronotus prope albifrons (102, 7 mm SL, NUP9621) b Rhamphichthys hahni (244, 4 mm SL, NUP9623) and c Brachyhypopomus gauderio (140, 9mm SL, NUP9622).
We obtained metaphase chromosomes from kidney cells using the air-drying technique proposed by
We used the C-banding method and staining with Giemsa after treatments with 0.1M HCl, Ba(OH)2 and 2×SSC solutions to analyse the distribution of heterochromatin, as described by
The probes used to detect 18S rDNA in the FISH analyses were obtained by amplified and cloned fragments of Oreochromis niloticus Linnaeus, 1758 (kindly provided by Dr Cesar Martins of the Universidade Estadual Paulista, Botucatu SP Brazil). We used the methods of
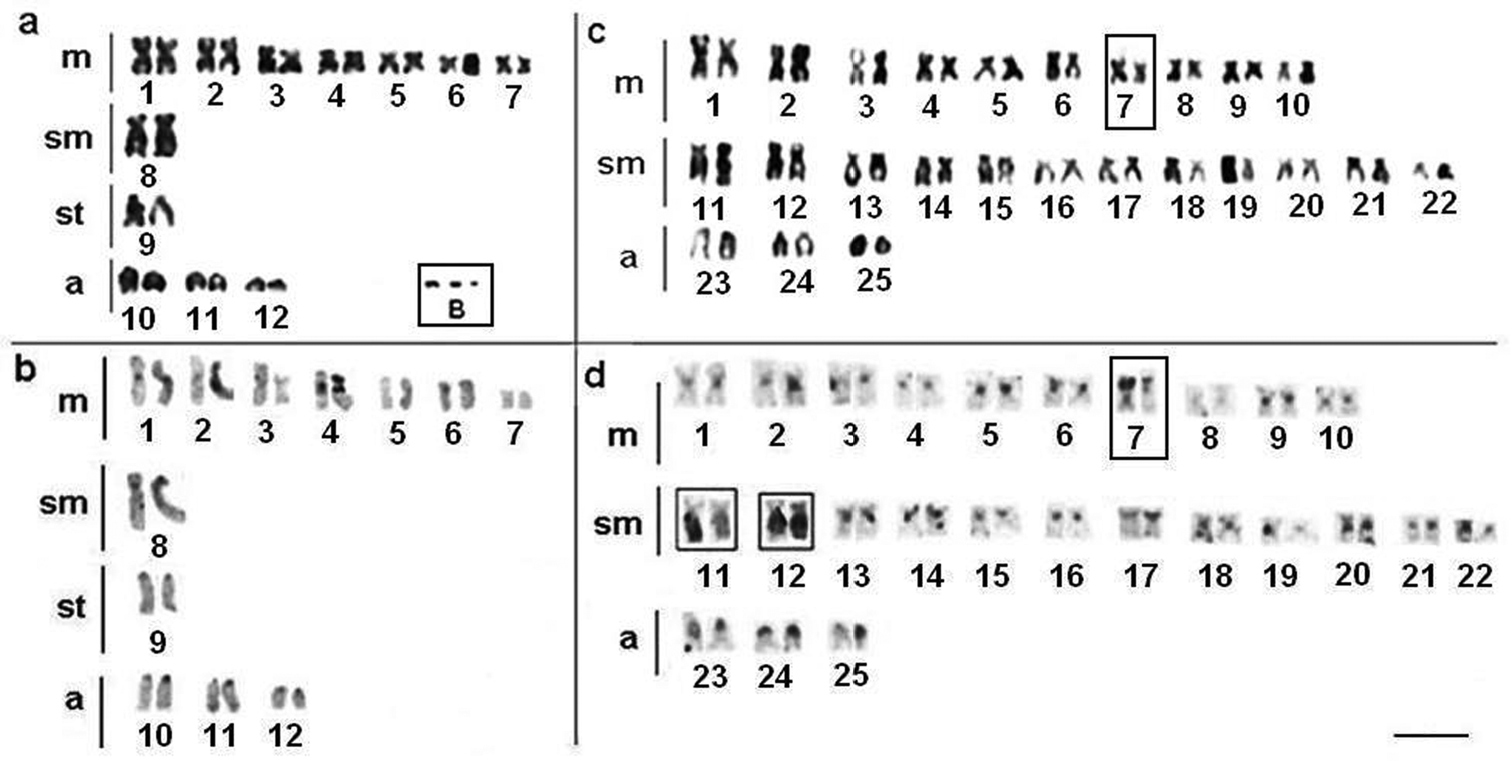
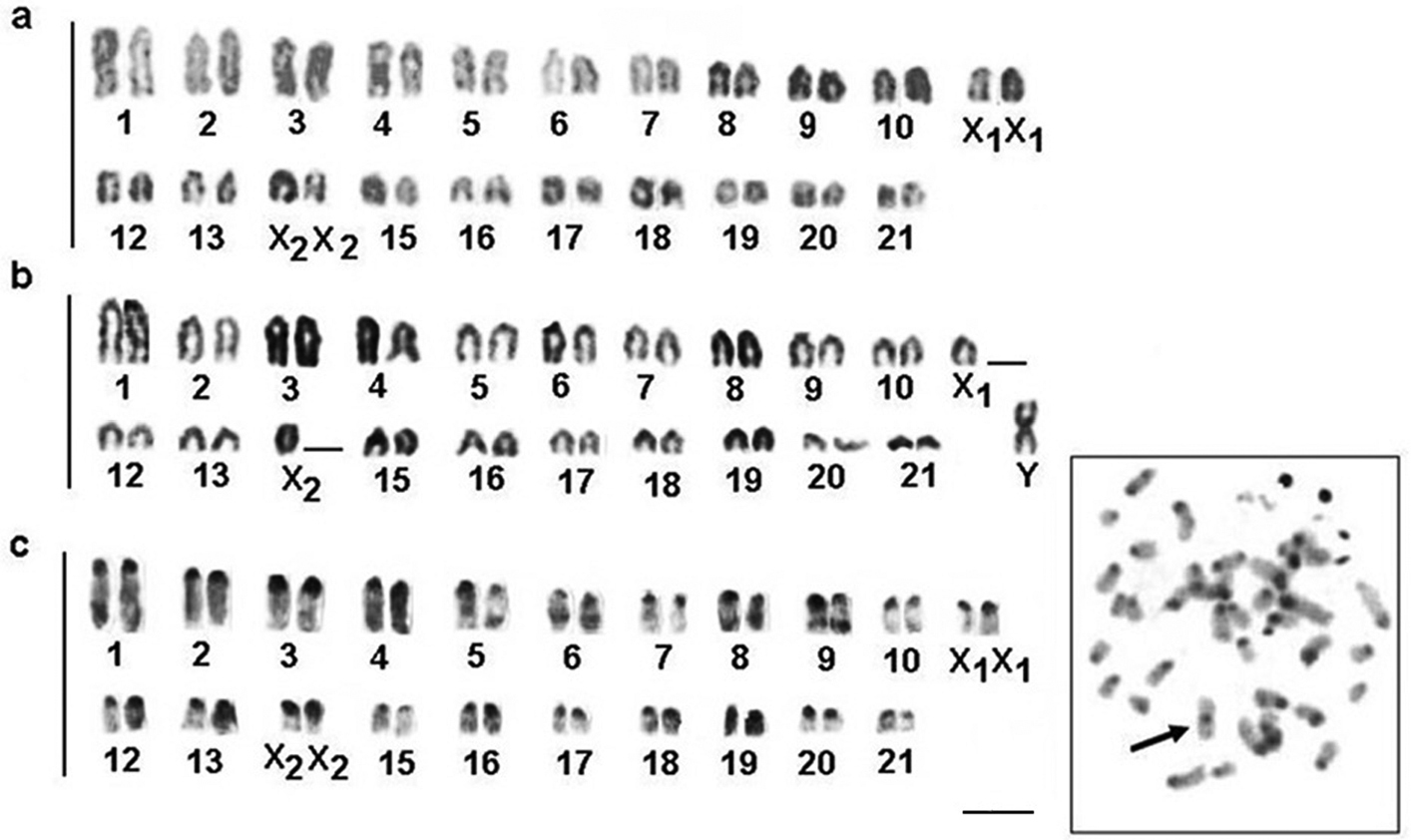
Apteronotus prope albifrons showed a diploid number of 24 chromosomes and a karyotype formula of 14m+2sm+2st+6a with a fundamental number (FN) of 42 (Fig. 3a). We observed constitutive heterochromatin distributed in small blocks throughout the pericentromere regions of most of the chromosomes (Fig. 3b) and in conspicuous blocks in the region adjacent to the nucleolar pair 4 secondary constriction (Fig. 3b). The Ag-NOR and 18S rDNA sites were located on the short arm of chromosome pair 4 (Fig. 4a, b), coinciding with the secondary constriction evident in some metaphases.
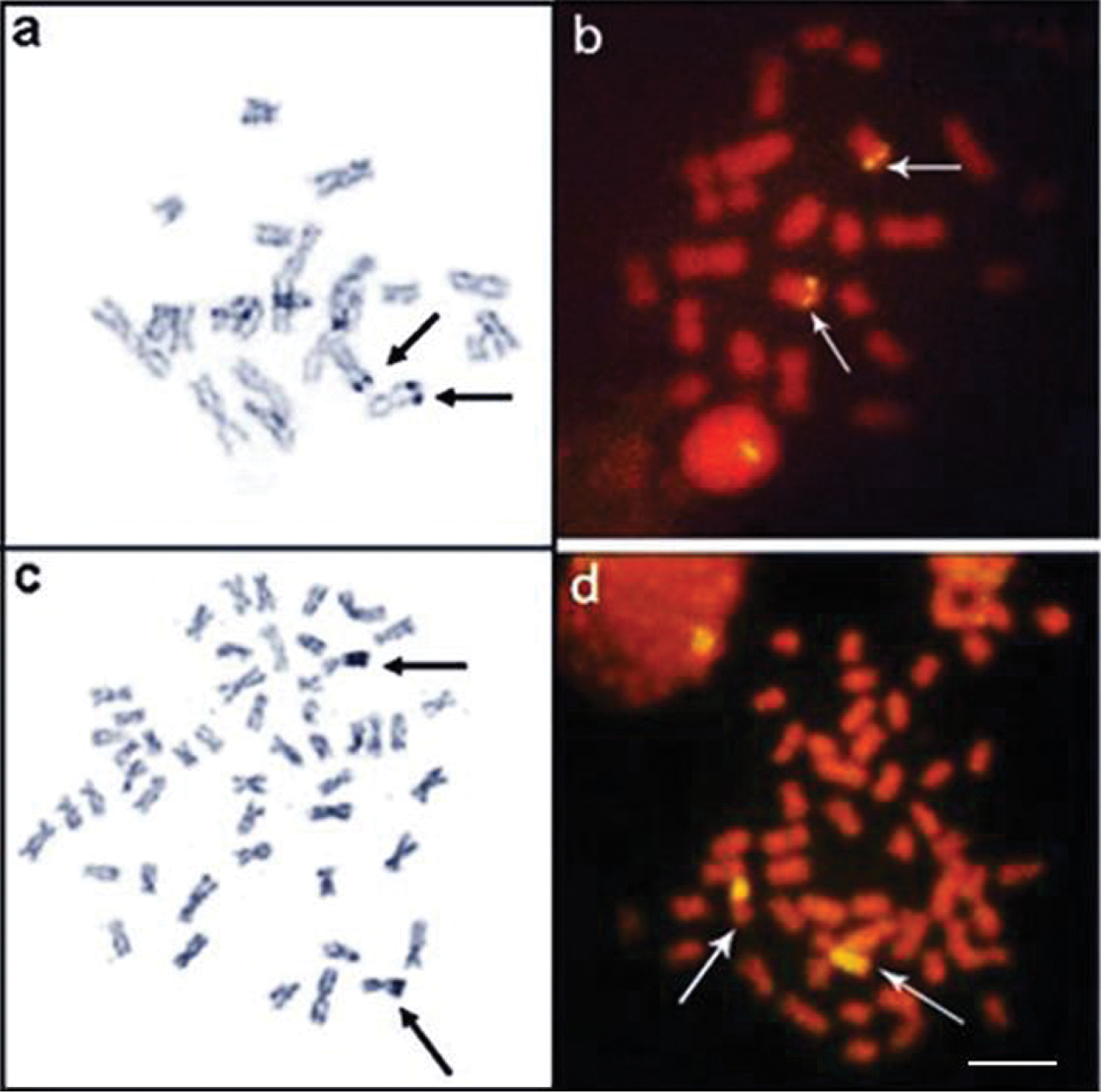
Karyotypes of Apteronotus prope albifrons and Rhamphichthys hahni after: Giemsa-staining a, c respectively and C-banding b, d; In evidence, B chromosomes of Apteronotus prope albifrons a and heterochromatic blocks adjacent to the nucleolar regions 4 b; c terminal secondary constriction on pair 7 and conspicuous heterochromatic regions on pairs 11 and 12 d in the karyotype of Rhamphichthys hahni. Bar = 5μm.
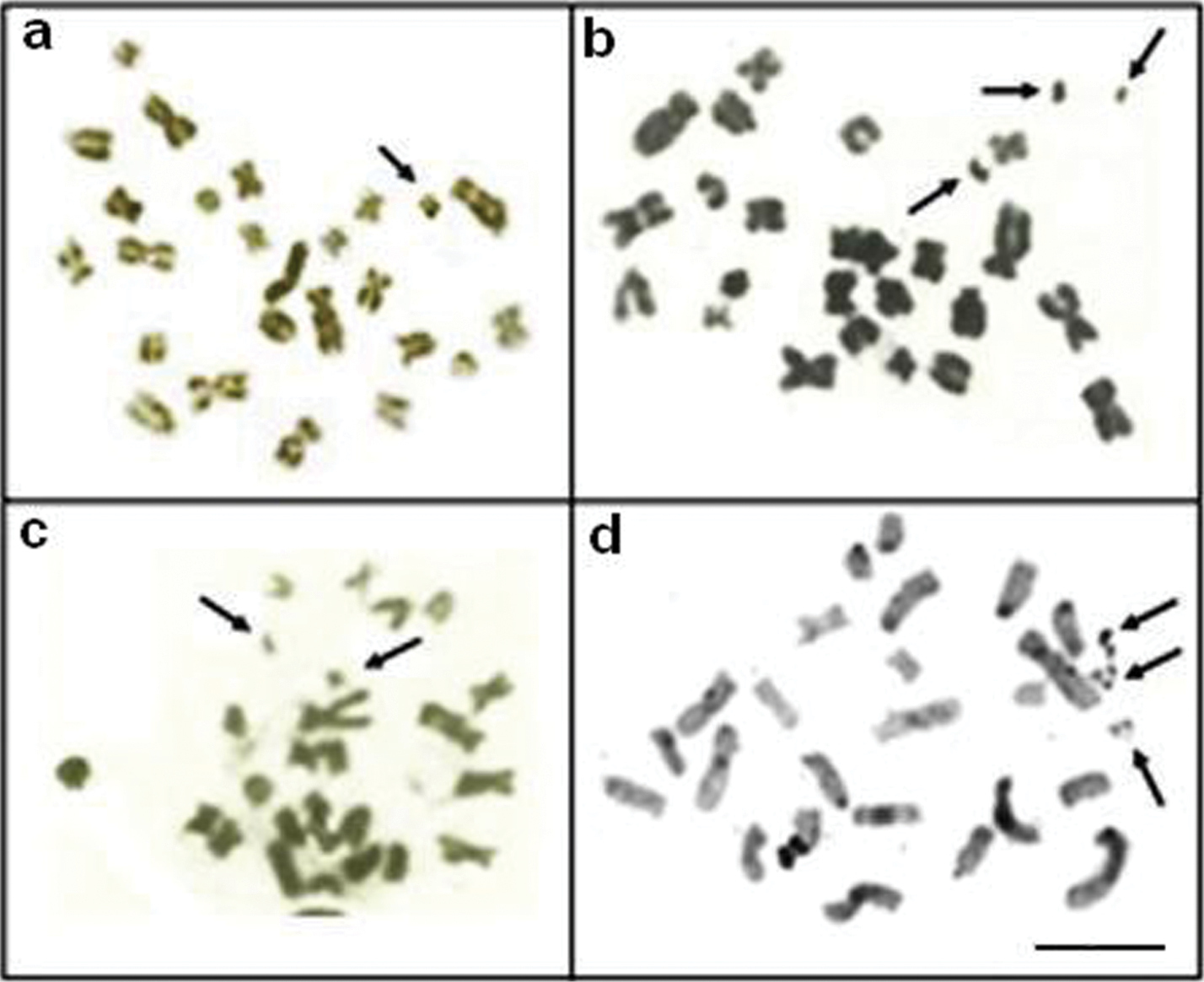
Metaphases showing Ag-NORs-bearing chromosomes and FISH using 18S rDNA probe of: Apteronotus prope albifrons a, b and Rhamphichthys hahni c, d Note an evident NOR region-sized heteromorphism of pair 7 of Rhamphichthys hahni c, d. Bar = 5μm.
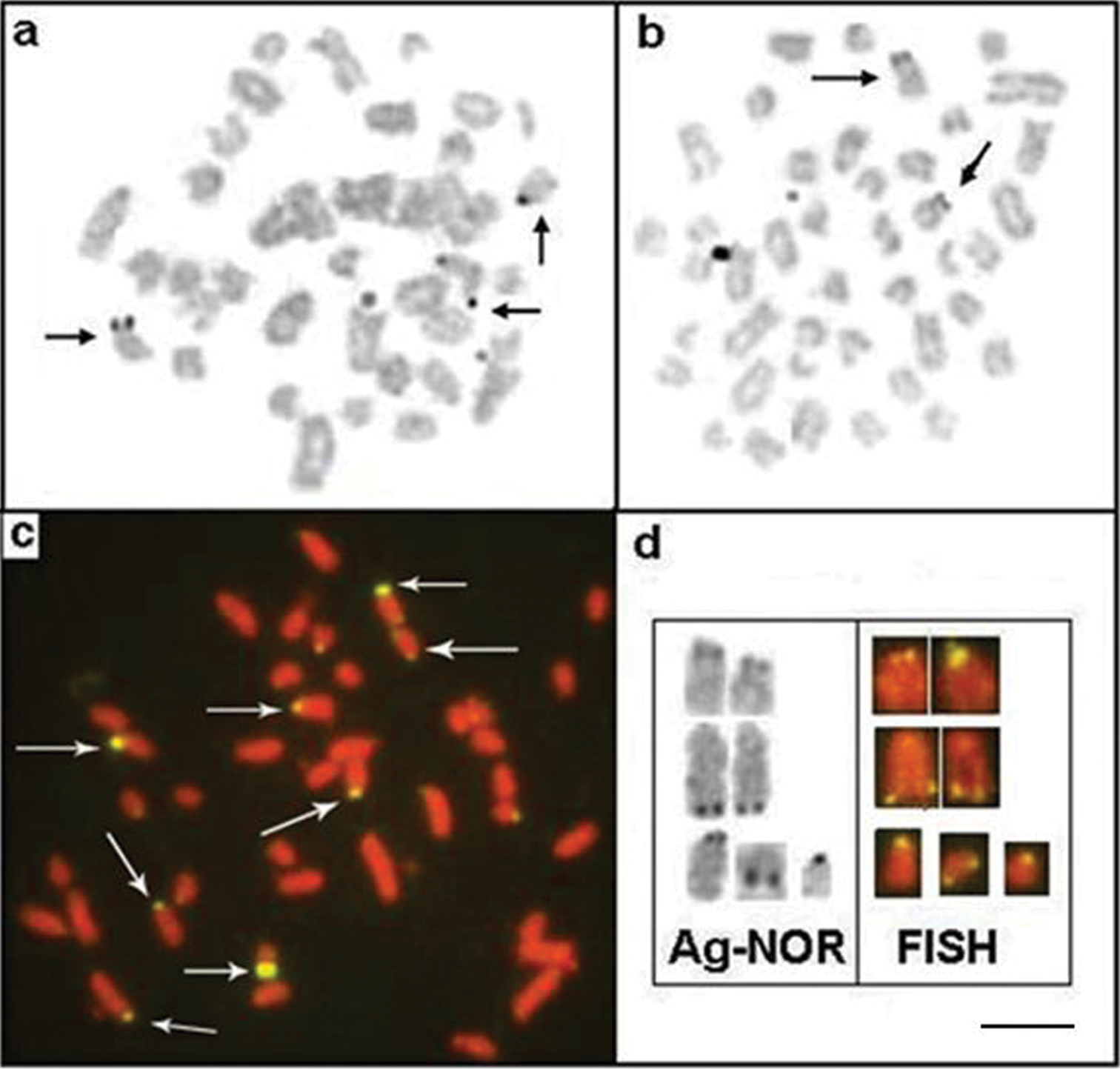
In addition to the normal chromosome complement, we found that three specimens of Apteronotus prope albifrons had one to three B microchromosomes in their somatic cells, with inter- and intra-individual variations (Fig. 3a, box and Fig. 5a–c). The B chromosomes showed no homology with the other chromosomes of the complement and, morphologically, these chromosomes were classified as acrocentric. We observed constitutive heterochromatin in the pericentromere and terminal position of the extra chromosomes (Fig. 5d).
Somaticmetaphases of Apteronotus prope albifrons stained with Giemsa a, b and c and C-banded d showing the B chromosomes (arrows). Bar = 5 μm.
Rhamphichthys hahni showed a diploid number of 50 chromosomes for both sexes and a karyotype formula of 20m+24sm+6a with a fundamental number (FN) of 94 (Fig. 3c). We observed constitutive heterochromatin in the pericentromere regions of most of the chromosomes and conspicuous blocks in three submetacentric chromosomes of the complement and also in the position close to secondary constriction of pair 7 (Fig. 3d). The Ag-NORs and 18S rDNA sites were located in the short arm of chromosome pair 7, coincideing with a terminal secondary constriction (Fig. 3c, box, and Fig. 4 c, d). We also observed an NOR region-sized heteromorphism in this pair (Fig. 4c, d).
Brachyhypopomus gauderio presents 2n=42 chromosomes in females and 2n=41 chromosomes in males. The female karyotype showed acrocentric chromosomes only (Fig. 6a) and the male karyotype showed 40 acrocentric chromosomes and one medium-sized metacentric chromosome (Fig. 6b). We found that the fundamental number for both sexes is 42. The difference in karyotype structure between the sexes demonstrates the presence of sex chromosomes, with a metacentric chromosome corresponding to a Y chromosome. Chromosomal pairs 11 and 14 are X1 and X2 in males and females, respectively. In fact, this condition characterizes a system of multiple sex chromosomes of the type X1X1X2X2/X1X2Y.
Karyotypes and metaphases of Brachyhypopomus gauderio after: Giemsa- staining in female a male b and C-banding in female c. Right, C-banded metaphase showing the Y chromosome (arrow). Bar = 5μm.
We found C-positive blocks (constitutive heterochromatin) in the pericentromere regions of all chromosomes in males and females (Figure 6c) of Brachyhypopomus gauderio, including the pericentromere region of the Y metacentric chromosome (Fig. 6, in the boxes). Furthermore, some chromosomes showed blocks of heterochomatin in the long arm in the interstitial and telomere positions. We observed Ag-NOR sites in the short and long arms of the acrocentric pairs (Fig. 7a, b), and the FISH technique revealed eight fluorescent signals (rDNA sites), including the sites stained by silver (Figure 7c). Figure 6d shows a correlation between some of the Ag-NOR and 18S rDNA sites.
Metaphases of Brachyhypopomus gauderio showing NORs regions: Ag-NORs-bearing chromosomes in the male a female b and eight 18S rDNA sites, arrows c; d correlation between some Ag-NOR and 18S rDNA regions. Bar = 5μm.
The karyotype structure we describe herein for Apteronotus prope albifrons (2n=24, 14m+2sm+2st+6a) is similar to the one reported by
Our report of B chromosomes in Apteronotus prope albifrons is the first description of these elements in the Gymnotiformes order. The mitotic instability presented by the B chromosomes of Apteronotus prope albifrons is likely due to their non-Mendelian behaviour during cell division, which is a common feature attributed to B chromosomes in other species.
The origin of B chromosomes is not yet well understood, although intraspecific origin of B chromosomes in some Neotropical fishes have been suggested as a result of the non-disjunction of an autosome, the formation of isochromosomes, centric fragments resulting from chromosomal rearrangements and amplifications of paracentric regions of a fragmented A chromosome (
Heterochromatinization plays an important role in the differentiation of B chromosomes. The B chromosomes are thought to be composed of repetitive sequences (heterochromatin) that lack protein-coding genes. The fact that the B chromosomes of Apteronotus prope albifrons appeared to be partially heterochromatic does not provide evidence of the presence or absence of coding genes. This would require a detailed molecular cytogenetic analysis with specific probes for FISH. Regarding the pattern of C bands of the other chromosomes of Apteronotus prope albifrons, we noted a similarity in the location of the heterochromatin when comparing it with the population studied by
The presence of only one NOR-bearing chromosome pair in Apteronotus prope albifrons in this study (confirmed by FISH) corresponds to the condition found in an Amazonian basin population of Apteronotus albifrons analysed by
Finally, we analysed the karyotypic data available so far for the Apteronotidae family and observed a relative numerical and structural variability, ranging from 2n=22 and 24 to 52 (Table 1), suggesting an evolutionary history of chromosomal rearrangements. This trend can also be seen when comparing the karyotypic formulas of the Apteronotus La Cépede, 1800 and Parapteronotus Albert, 2001 genera (Table 1). However, the absence of cytogenetic data for other species of the Apteronotidae family makes it difficult to understand the karyotypic interrelationships and the types of rearrangements that have resulted in the diploid values in this group so far. In addition, cytogenetic information for other species of this family would be helpful for clarifying the origin of the B chromosomes discussed herein.
The karyotype structure of Rhamphichthys hani and Brachyhypopomus gauderioCytogenetic studies in the Rhamphichthyidae family are still scarce and the karyotype of Rhamphichthys hahni is reported herein for the first time. The cytogenetic data available for Rhamphichthysi Müller et Troschel, 1848show less variation in diploid number and chromosome structure (see Table 1). For example, the comparison of the karyotype of Rhamphichthys rostratus Linnaeus, 1766 (
The presence of NOR in only one pair of chromosomes found in Rhamphichthys hahni coincides with the patterns observed in Rhamphichthys marmoratus Castelnau, 1855 and Rhamphichthys rostratus (
The C-band pattern in Rhamphichthys hahni is similar to the heterochromatin location in the pericentromere region in many species of Gymnotiformes. The NOR region is associated with heterochromatin, which is frequently found in many fish species. However, three medium-sized submetacentric chromosomes show conspicuous heterochromatic blocks on the long arm. The distribution and amount of heterochromatin may have an important evolutionary role in the chromosomes of many fish species, including sex chromosomes, as reported in Eigenmannia virescens (
The chromosome formula discovered by us in Brachyhypopomus gauderio corresponds to the data previously reported by
The origin of the multiple sex determination system, X1X1X2X2/X1X2Y in Brachyhypopomus gauderio (cited as Brachyhypopomus pinnicaudatus) was discussed by
The detection of up to eight chromosomes with a fluorescent signal (18S rDNA sites) indicates multiple NOR systems in Brachyhypopomus gauderio. Furthermore, the difference in the results between the methodologies (Ag-NOR and FISH) suggests that not all ribosomal DNA sites were active in the previous interphase. Also, chromosomal rearrangements such as translocations and/or transpositions, resulting in the dispersion of ribosomal genes, might explain this variability. No information is provided on the sites of NORs in the Brachyhypopomus pinnicaudatus (currently Brachyhypopomus gauderio) population analysed by
The authors are grateful to Dr Carla Simone Pavanelli (Nupelia, UEM) for identifying the species and to the Brazilians agencies, CAPES - Universidade Estadual de Maringá (Maringá, Parana state) for their financial support.