(C) 2012 Anderson José Baia Gomes. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
The family Phyllostomidae belongs to the most abundant and diverse group of bats in the Neotropics with more morphological traits variation at the family level than any other group within mammals. In this work, we present data of chromosome banding (G, C and Ag-NOR) and Fluorescence In Situ Hybridization (FISH) for representatives of Rhinophylla pumilio Peters, 1865 collected in four states of Brazil (Amazonas, Bahia, Mato Grosso and Pará). Two karyomorphs were found in this species: 2n=34, FN=64 in populations from western Pará and Mato Grosso states and 2n=34, FN=62 from Amazonas, Bahia, and northeastern Pará and Marajó Island (northern). Difference in the Fundamental Number is determined by variation in the size of the Nucleolar Organizer Region (NOR) accompanied with heterochromatin on chromosomes of pair 16 or, alternatively, a pericentric inversion. The C-banding technique detected constitutive heterochromatin in the centromeric regions of all chromosomes and on the distal part of the long arm of pair 15 of specimens from all localities. FISH with a DNA telomeric probe did not show any interstitial sequence, and an 18S rDNA probe and silver staining revealed the presence of NOR in the long arm of the pair 15, associated with heterochromatin, and in the short arm of the pair 16 for all specimens. The intra-specific analysis using chromosome banding did not show any significant difference between the samples. The comparative analyses using G-banding have shown that nearly all chromosomes of Rhinophylla pumilio were conserved in the chromosome complements of Glossophaga soricina Pallas, 1766, Phyllostomus hastatus Pallas, 1767, Phyllostomus discolor Wagner, 1843 and Mimon crenulatum Geoffroy, 1801, with a single chromosomal pair unique to Rhinophylla pumilio (pair 15). However, two chromosomes of Mimon crenulatum are polymorphic for two independent pericentric inversions. The karyotype with 2n=34, NF=62 is probably the ancestral one for the other karyotypes described for Rhinophylla pumilio.
Biodiversity, Amazon rainforest, Chiroptera, cytogenetics
Traditionally, the subfamily Carolliinae (sensu
Cytogenetic studies in Carolliinae have shown different rates of chromosomal evolution between both genera. The genus Carollia has two karyomorphs: 2n=20/21 with a multiple sex chromosome system (XX/XY1Y2), observed in most species (
Cytogenetic samples of Rhinophylla pumilio from different localities. Numbers of sites correspond to numbers of triangles on the map (Fig. 1).
| Site | n | Locality/State | 2N/FN | Methods | Geographical coordinates |
|---|---|---|---|---|---|
| 1 | 1♂+1♀ | Chaves, Pará | 34/62 | G | 00°24'55.3"S; 49°58'44.1"W |
| 1 | 3♀ | 34/62 | |||
| 2 | 1♂ | Marituba, Pará | 34/62 | G, C | 01°16'37.5"S; 48°20'14.9"W |
| 3 | 1♂ | Belém, Pará | 34/62 | G, C, NOR, Telomere, rDNA, CMA3 | 01°13'29.3"S; 48°32'59.0"W |
| 3 | 1♂ | 34/62 | G, C | ||
| 4 | 1♂+1♀ | Santa Barbara, Pará | 34/62 | G | 01°13'57.4"S; 48°16'34.4"W |
| 4 | 4♂+2♀ | 34/62 | |||
| 5 | 1♀ | Capanema, Pará | 34/62 | C | 01°24'02.5"S; 48°29'02.4"W |
| 6 | 1♂ | Peixe-Boi, Pará | 34/62 | G, C | 01°11'11.0"S; 47°19'28.5"W |
| 6 | 1♂ | 34/62 | G, C, rDNA, CMA3 | ||
| 7 | 2♂+1♀ | Oriximiná, Pará | 34/62 | G, C | 01°39'03.3"S; 56°20'30.6"W |
| 8 | 1♀ | Faro, Pará | 34/62 | G, C | 02°03'53.1"S; 56°37'57.4"W |
| 9 | 1♂ | Juruti, Pará | 34/64 | G, C, NOR, rDNA | 02°29'38.8"S; 56°11'27.1"W |
| 9 | 1♀ | 34/64 | G, C, rDNA | ||
| 10 | 1♀ | Itaituba, Pará | 34/64 | 04°16'26.6"S; 55°56'47.6"W | |
| 10 | 1♂ | 34/64 | G, C, rDNA, CMA3 | ||
| 11 | 1♂+1♀ | Itaituba, Pará | 34/64 | G, C | 04°28'20.5"S; 56°17'03.7"W |
| 12 | 1♂+3♀ | Itacoatiara, Amazonas | 34/62 | G, C | 02°58'49.6"S; 58°57'51.0"W |
| 12 | 1♀ | 34/62 | |||
| 13 | 1♂+4♀ | Potriguaçú, Mato Grosso | 34/64 | G, C | 09°51'53.7"S; 58°13'06.8"W |
| 14 | 1♂ | Ilhéus, Bahia | 34/62 | G, C, NOR | 14°47'52.0"S; 39°10'15.0"W |
Previous cytogenetic studies on Rhinophylla pumilio. Numbers of sites correspond to numbers of squares on the map (Fig. 1).
| Site | Region | Geographical coordinates | 2n/FN | References |
|---|---|---|---|---|
| 1 | Suriname | 05°27'00"S; 55°12'00"W | 34/64 | Honeycutt et al. 1980, |
| 2 | Suriname | 03°46'00"S; 56°10'00"W | 34/56 |
|
| 3 | Colombia | 04°07'43"S; 69°56'37"W | 36/62 |
|
| 4 | Brazil-Bahia | 14°17'29"S; 39°51'18"W | 26/48 |
|
The monophyly of the subfamily Carolliinae and the sister-group relationships of Carollia and Rhinophylla have been supported by a phylogenetic analysis based on morphological data (
Therefore, we analyzed, through conventional cytogenetic (G-, C- banding and Ag-NOR staining) techniques and Fluorescence In Situ Hybridization (FISH) with rDNA and Telomere probes, two karyotypes of Rhinophylla pumilio and discussed the biogeographical chromosome variation by comparing karyotypes of this species with representatives of two subfamilies of Phyllostomidae (Glossophaginae and Phyllostominae).
Material and methods Specimens analyzedCytogenetic preparations of Rhinophylla pumilio were obtained from 40 specimens collected in four states in Brazil: Pará state – 16 males and 13 females, Amazonas state – 1 male and 4 females, Mato Grosso state – 1 male and 4 females, Bahia state – 1 male (Fig. 1, Table 1). The bats were collected in the field using mist nets during the expeditions to faunal inventories. Comparative cytogenetic analyses were performed with Glossophaga soricina Pallas, 1766 (from Santa Barbara), Phyllostomus hastatus Pallas, 1767 (from Peixe-Boi), Phyllostomus discolor Wagner, 1843 (from Belém) and Mimon crenulatum Geoffroy, 1801 (from Faro). Chromosomal preparations and tissue biopsies were sent to the Cytogenetics Laboratory at Universidade Federal do Pará. Animals were fixed in 10% formalin preserved in 70% ethanol and deposited in the mammal’s collection of the Museum Paraense Emilio Goeldi, mammal’s collection of the Santa Cruz State University, Ilhéus-Bahia, Zoology Museum of the Mato Grosso Federal University and Zoology Museum of the West Pará Federal University.
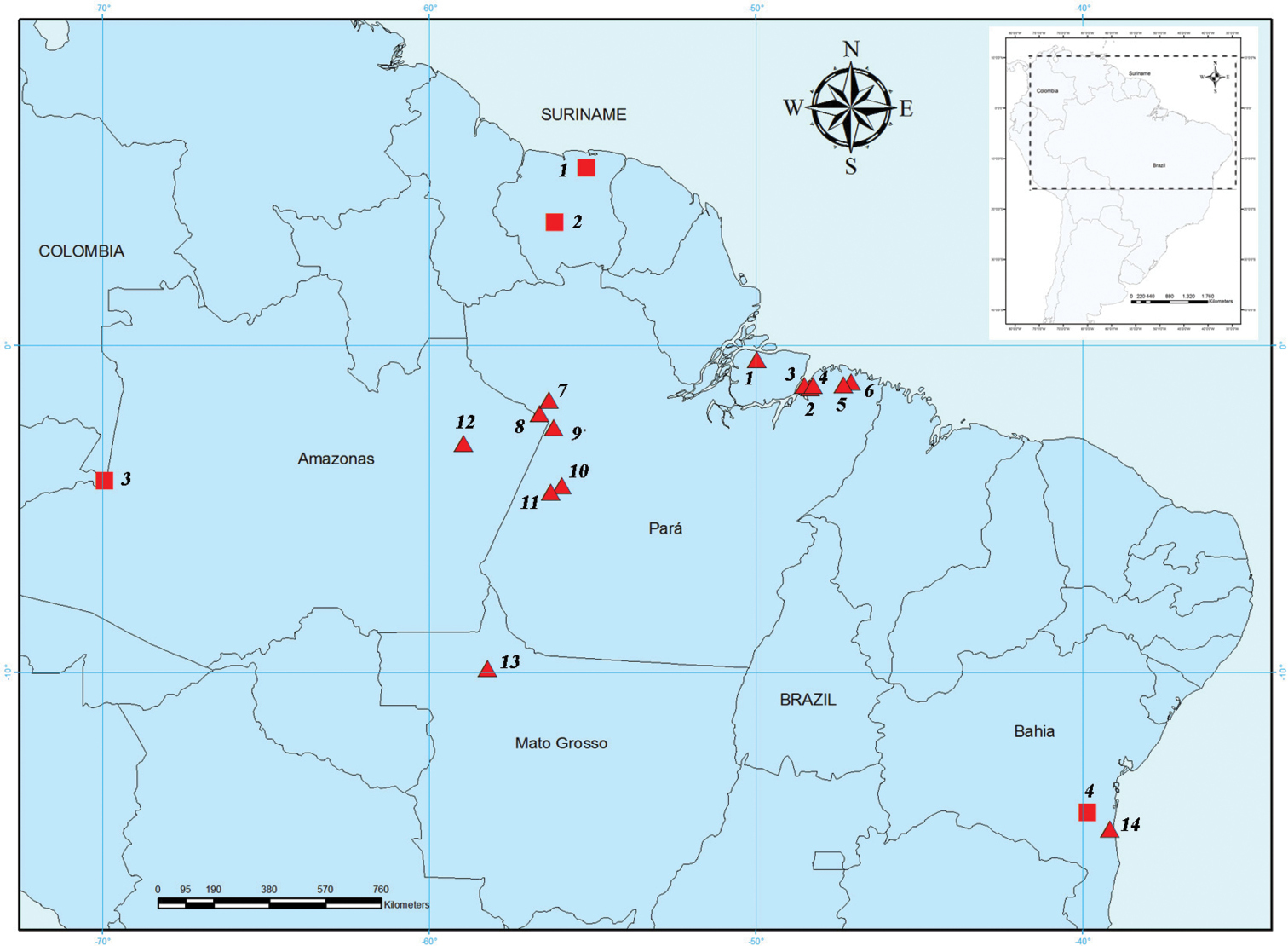
Map of collected samples of Rhinophylla pumilio. Squares indicate the sites from where previous cytogenetic descriptions were performed whereas triangles represent the cytogenetic samples studied herein (see Tables 1 and 2 for locality details). Numbers of sites correspond to numbers on Tables 1 and 2.
The chromosome spreads were obtained from bone marrow following
Fluorescence In Situ Hybridization using digoxigenin-labeled telomeric probes (All Human Telomere Probes, Oncor) was performed according to the manufacturer’s protocol. To confirm the position of the NORs, 18S rDNA probes were amplified by BACs (Bacterial Artificial Chromosomes), labeled by nick translation and subsequently detected with avidin-Cy3 or anti-digoxigenin- FITC. Briefly, the slides were incubated in RNAse and pepsin solutions following
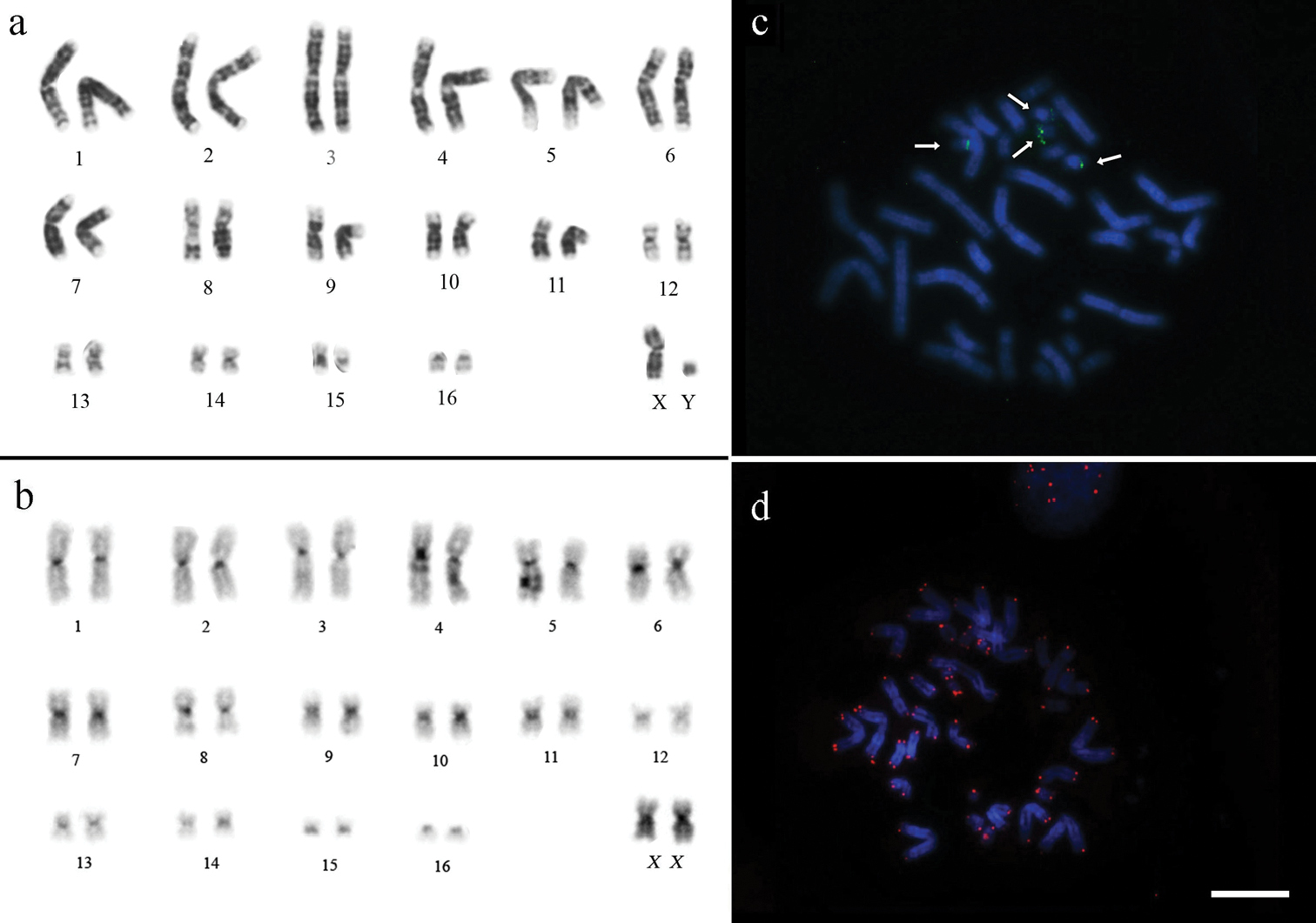
All studied specimens of Rhinophylla pumilio have the same chromosome number – 2n=34. The autosomal complement consists of 15 pairs biarmed (metacentric and submetacentric) and one pair of acrocentric chromosomes (pair 16) in samples collected from Bahia, Amazonas, northeastern Pará and Marajó Island (north of Para) (Fig. 2a). In contrast, the chromosome pair 16 of specimens from west Pará and Mato Grosso is biarmed (Fig. 3a). The X chromosome is a medium-sized metacentric chromosome and the Y is a small acrocentric.
Karyotypes of Rhinophylla pumilio from northeastern Pará (except C-banding obtained from specimens from Amazonas state) a G-banding b C-banding c 18S rDNA FISH and d telomeric FISH. Arrows show NORs in the chromosome pairs 15 and 16. Bar = 10 µm.
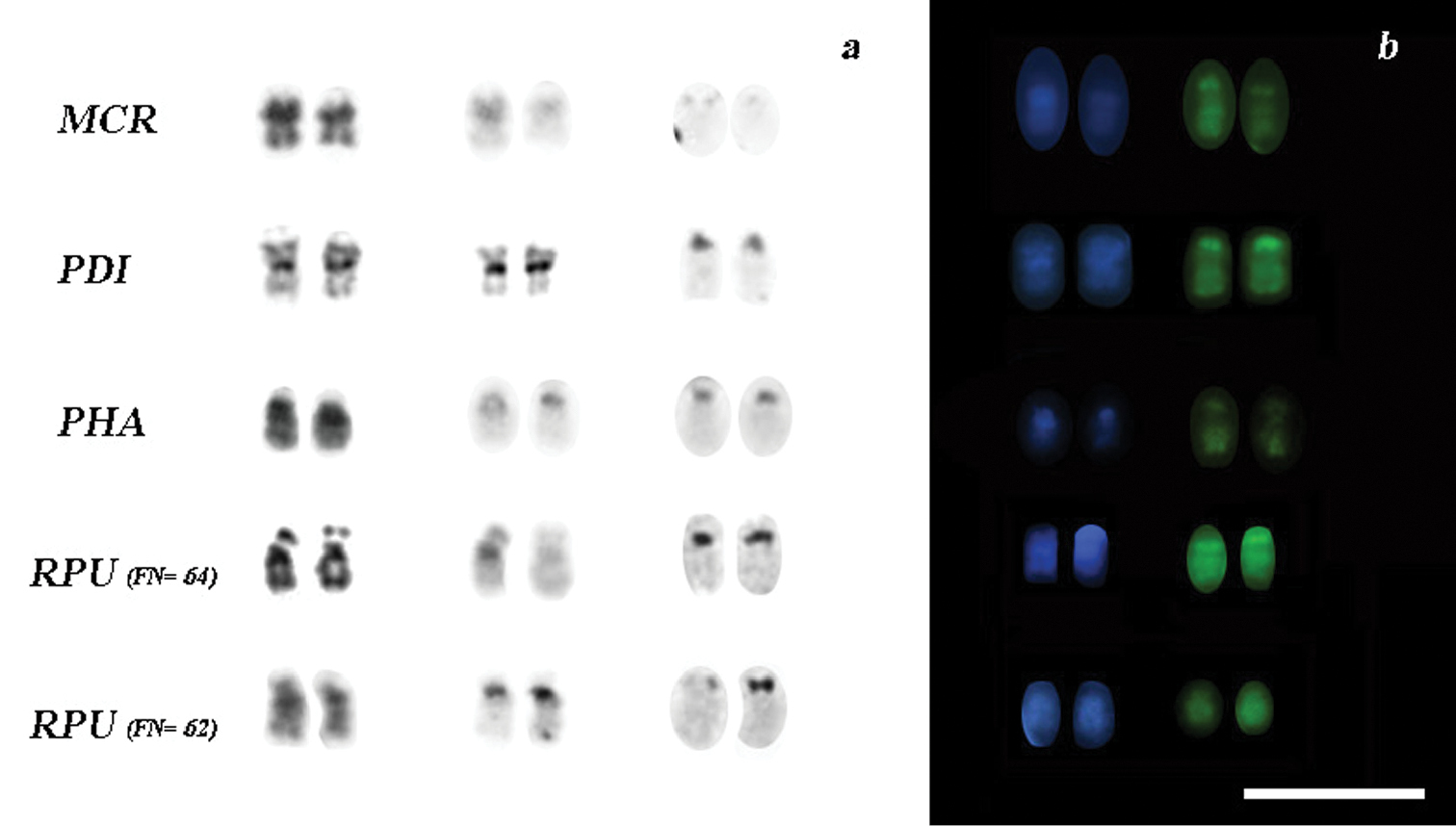
Variation of chromosome pair 15 (16 in Rhinophylla pumilio) in the analyzed species a chromosomes after G, C and Ag-NOR sequential staining b patterns of double staining with DAPI-CMA3. Bar = 10 µm.
The constitutive heterochromatin was found in the centromeric regions of all chromosomes and at the distal part of the long arm of pair 15 for all specimens (Fig. 2b). Telomere sequences were observed at the tips of chromosomes (Fig. 2d). The rDNA probes and staining with silver nitrate confirmed the presence of NORs in the long arm of the pair 15 and short arm of the pair 16 (Fig. 2c). The FISH with rDNA and subsequent double staining with DAPI and CMA3 are in agreement with the patterns of G-bands and R-bands, respectively, where the R-bands show the tips of the chromosomes and its association with the NOR (Fig. 3b).
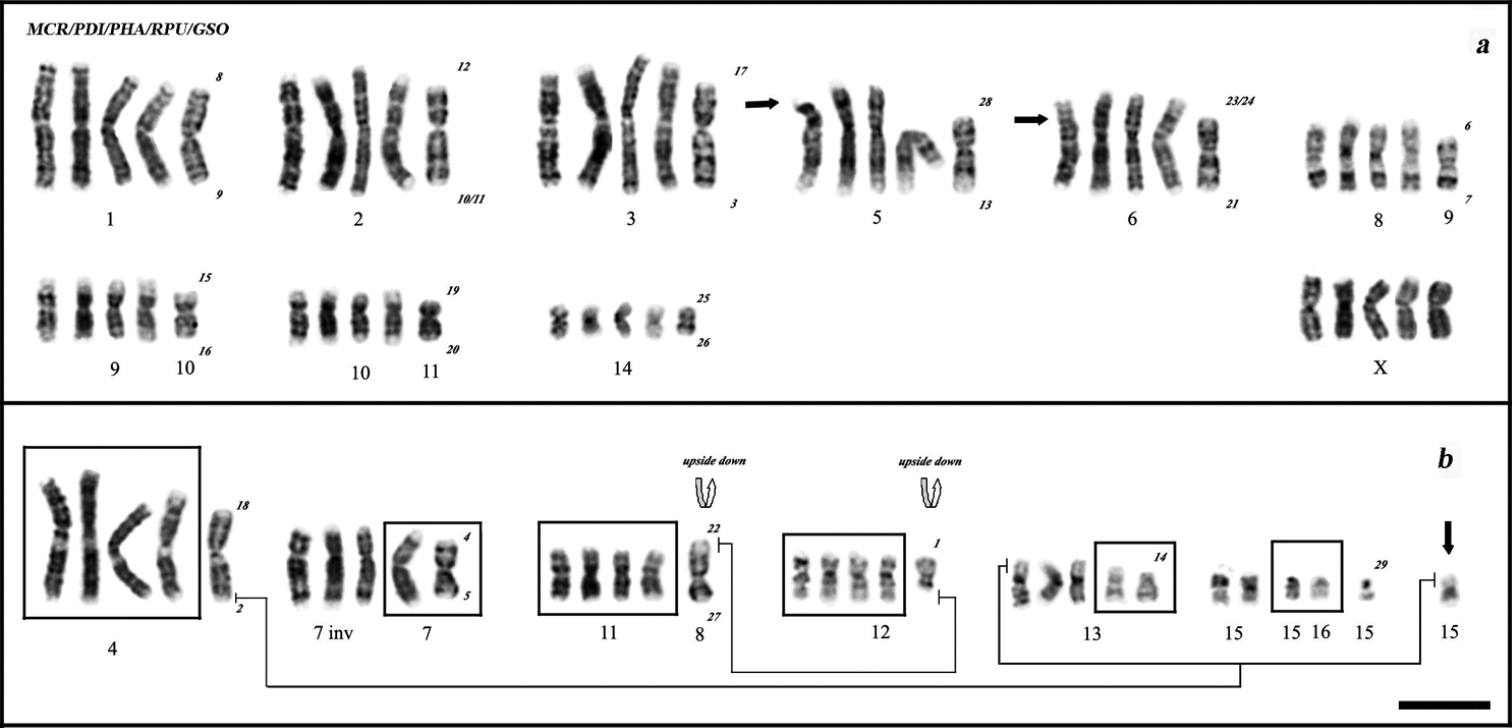
The comparative analysis with Phyllostomus hastatus, Phyllostomus discolor, Mimon crenulatum (Phyllostominae) and Glossophaga soricina (Glossophaginae) (Fig. 4a) suggests that the karyotypes of Rhinophylla pumilio here described have nearly all chromosome pairs shared with these species, although one pair was autapomorphic to Rhinophylla pumilio (Fig. 4b). Analyzed species are different in the number of chromosomes (34 in Rhinophylla pumilio and 32 in other species) and the fundamental number (58 in Phyllostomus hastatus, 60 in Mimon crenulatum, Phyllostomus discolor, Glossophaga soricina and 62/64 in Rhinophylla pumilio). The heterochromatin presents in the centromeric regions of all species with additional blocks in the short and long arms of the 15th pair of Mimon crenulatum and Glossophaga soricina, respectively. Chromosomes of 5th and 6th pairs of Mimon crenulatum exhibit two polymorphic conditions derived probably from pericentric inversions that could cause the acrocentric and subtelocentric forms, respectively. Both specimens are heterozygous for 6th pair and homozygous for normal and rearranged forms of 5th chromosome pair. The NORs in this species are localized in the short arm of 15th pair and in the Y chromosome.
Comparative analysis using G-banded chromosomes of Mimon crenulatum, Phyllostomus discolor, Phyllostomus hastatus, Rhinophylla pumilio and Glossophaga soricina, from left to right a Conserved chromosomes among species, arrows show the centromeric position in Mimon crenulatum b Chromosomal differences among species. Black arrow indicates autapomorphic chromosome in Rhinophylla pumilio. Numbers (beside Glossophaga soricina) correspond to the chromosomal nomenclature applied to arms of Macrotus waterhousii in Glossophaga soricina according to
Our G-, C-, and Ag-NOR banding analyses have shown two distinct karyotypes for specimens of Rhinophylla pumilio from localities ranging more than 1000 km. The differences between these karyotypes may be caused by a pericentric inversion in the chromosome pair 16 or, alternatively, an amplification of rDNA cistrons accompanied with a faint block of heterochromatin in Rhinophylla pumilio with FN=64 (Fig. 3a). This segment is coincident with CMA3 positive staining for NOR and DAPI positive to the heterochromatic block (Fig. 3b).
Comparative analysis of karyotypes from different geographic localities (Table 2) allows discussing the morphology and number of chromosomes. Since only data of conventional staining or karyotype formula were described in the literature we had to restrict our comparisons to number and basic morphology of chromosomes. In this way, specimens of Rhinophylla pumilio collected on the Marajó island and northeastern Pará (Fig. 1, triangles 1, 2, 3, 4, 5, and 6) in the left side of the Amazon basin on Pará and Amazonas (triangles 7, 8 and 12) and Bahia (triangle 14) have 2n=34 and FN=62. Meanwhile, the samples from western Pará (triangles 9, 10 and 11) and Mato Grosso (triangle 13) presented the same fundamental number as specimens collected from Suriname, with 2n=34, FN=64 (
Karyotype with 2n=26 and FN=48 described by
Another cytogenetic study on specimens of Rhinophylla pumilio from Colombia described a karyotype with 2n=36 and FN=62, (
Comparative analysis of chromosome banding patterns of Rhinophylla pumilio was undertaken with representatives of two other subfamilies of Phyllostomidae bats: Phyllostomus hastatus, Phyllostomus discolor, Mimon crenulatum (Phyllostominae) and Glossophaga soricina (Glossophaginae). Karyotypes of these species supposed to be ancestral for their respective subfamilies (
Comparative analysis revealed that there are an extensive number of conserved chromosomes shared among these species. However, Rhinophylla pumilio shared more characters with Phyllostominae species than Glossophaga soricina (Fig. 4b). Based on outgroup comparisons,
Furthermore, other differences among karyotypes (Fig. 4b) are a pericentric inversion on pair 7 of Phyllostomus hastatus (
Another interesting problem in our comparative analysis is the pair 16 in Rhinophylla pumilio, which has two chromosomal traits similar to those observed within representatives of genus Phyllostomus Lacépède, 1799. The difference between the karyotypes of Phyllostomus hastatus and Phyllostomus discolor consists of a pericentric inversion of the pair 15 (
Among species of genus Carollia karyotypes are highly rearranged and after the reciprocal chromosome painting
Finally, we believe that variation of karyotypes along the area of Rhinophylla pumilio is correlated with intraspecific variation where the karyomorphs would be derived from ancestral karyotype with 2n=34, FN=62, since this karyotype is similar to other close related species at the chromosome level. However, additional analyses will be necessary to elucidate the biogeographical patterns related to the chromosome variation in Rhinophylla pumilio.
We thank to Manoel Rodrigues (Juris Ambientes Consultores), Flávio Eduardo Pimenta and Ana lima (Aotus Consultoria), Pablo Suarez, Dionísio Pimentel, Heriberto Figueria and the staff of CNEC- Juruti for Help us in the field expedition in the area of Alcoa’s Juruti Bauxite mine. We thank to Biodinamica-Rio, Aotus Consultoria and Talita Ribas, Eloiza Soarez, Fabio Sarmento, Fabio Augustos and Ramon Araujo for help us in the bats expedition in the Electric power transmission from Oriximina (Pará state) to Silves (Amazonas state) and Thayse Benathar for the chromosome preparation from Mato Grosso state and in the area TERFRON project on Itaituba. We are grateful to Cibele Sotero-Caio for a critical analysis of this work. This research was supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Conselho Aperfeiçoamento de Pessoal de Nível Superior), UFOPa (Universidade Federal do Oeste do Pará) and UFPa (Universidade Federal do Pará).