(C) 2012 Josiane Baccarin Traldi. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
In this study, which is the first karyotype analysis of Hypostomus iheringii, nine specimens collected in Córrego da Lapa (tributary of the Passa-Cinco River) showed a diploid number of 80 chromosomes. Silver nitrate staining and fluorescence in situ hybridization (FISH) with an 18S rDNA probe revealed the presence of multiple nucleolus organizer regions (NORs) (chromosome pairs 13, 20, and 34). FISH with a 5S rDNA probe showed that this cistron was only present in chromosome pair 2. When the karyotypes of individual animals were compared, unique heterochromatic polymorphisms were detected on chromosome pairs 1 and 5. Specifically, specimens had heterochromatic blocks (h+h+) on both chromosomes, one chromosome with heterochromatic blocks (h+h-) or chromosomes that lacked heterochromatic blocks (h-h-). Considering that heteromorphic pattern is not correlated with variation in size, the process of heterochromatinization might act on the long arms of these chromosomes. In summary, all chromosomal markers indicate that the karyotype of Hypostomus iheringii is highly differentiated and that the heterochromatinization of chromosomal segments may have contributed to its karyotypic differentiation.
chromosome variation, fish, heterochromatinization, Hypostomini
Loricariidae is a speciose group of Neotropical fishes that is composed of six subfamilies: Hypoptopomatinae, Hypostominae, Lithogeneinae, Loricariinae, Neoplecostominae, and Delturinae (
Cytogenetic studies in Hypostomus indicate that there is great variability in various karyotypic aspects, which contributes to enormous complexity of the group (
The amplification and mobility of heterochromatic blocks of chromosomes are well documented in some organisms (Hamilton et al. 1990, Modi 1993). Sequences of satellite DNA appear to play an important role in the evolution of the genome of different organisms by promoting chromosome rearrangements and exhibiting rapid differentiation due to intragenomic mobility taking important role in karyotype evolution and speciation due to gene flow restriction (
In this study, the first karyotype analysis of Hypostomus iheringii (Regan, 1908) was performed using classic (Giemsa staining, C-banding, and Ag-NOR) and molecular (fluorescence in situ hybridization - FISH) cytogenetic methods, emphasizing the distribution of heterochromatic blocks, interrelating and discussing the possible role of heterochromatin in the diversification of the genomes of Loricariidae.
Materials and methods Animals and mitotic chromosome preparationsNine specimens (five males and four females) of Hypostomus iheringii from the Córrego da Lapa, a tributary of the Passa-Cinco River in Ipeúna, São Paulo, Brazil, were analyzed. These specimens were deposited in the Museum of Zoology at the University of São Paulo, under voucher number MZUSP 106769. The animals were anesthetized with clove oil, according to the method described by
The chromosomes were stained with a solution of 5% Giemsa. C-banding was performed following the protocol described by
The physical mapping of 18S and 5S rDNA on the chromosomes was obtained by FISH, as described by
The chromosomes were classified as metacentric (m), submetacentric (sm), subtelocentric (st), and acrocentric (a), according to the ratio of arms proposed by
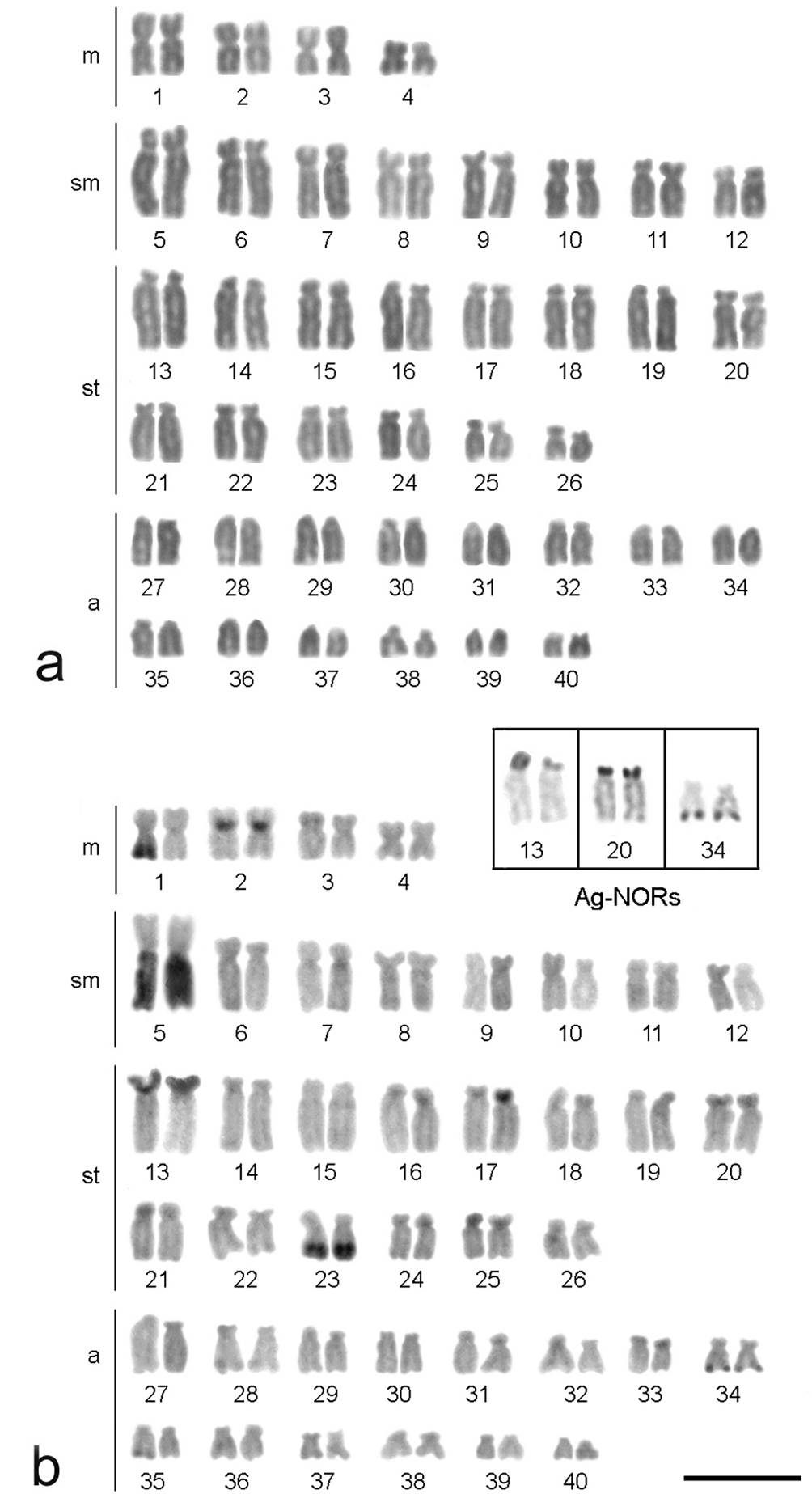
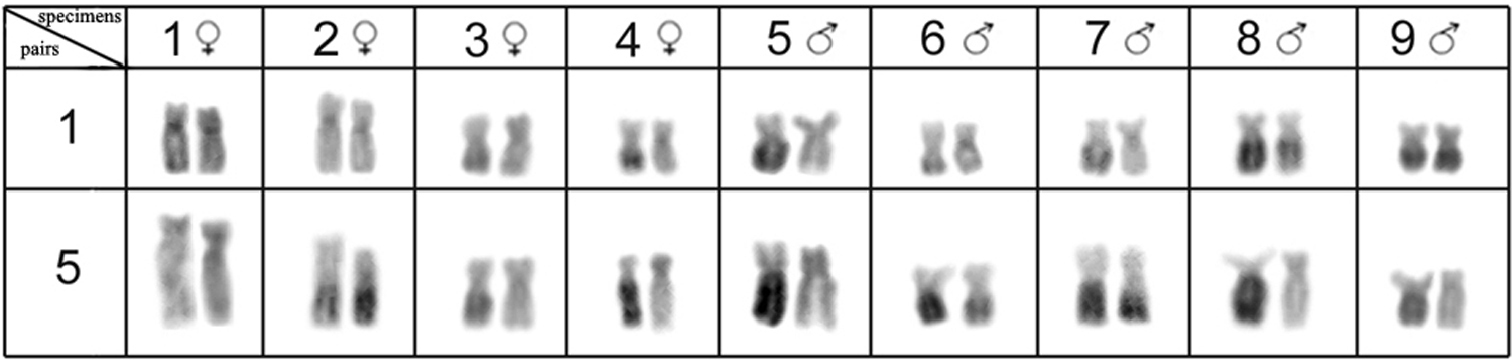
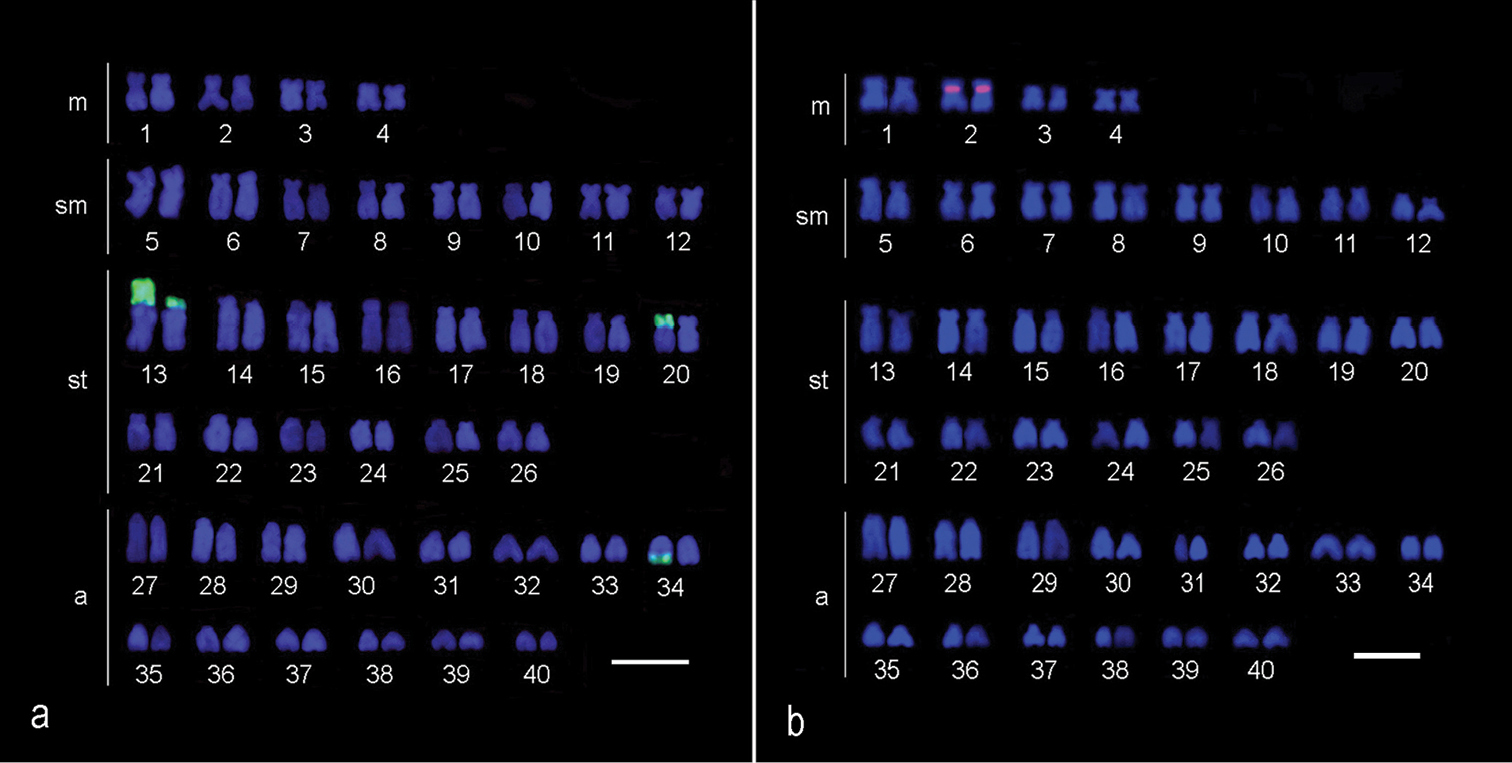
All of the Hypostomus iheringii specimens showed a diploid number of 80 chromosomes (8m + 16sm + 28st + 28a, NF=132) without a heteromorphic sexual system (Fig. 1a). C-banding analysis revealed a small section of heterochromatin, preferentially located in the terminal portions of some of the chromosomes (Fig. 1b). Two conspicuous heterochromatic blocks were found in the terminal position of the long arm of chromosome pairs 1 (m) and 5 (sm) with inter-individual variations. Specifically, specimens had heterochromatic blocks (h+h+) on both chromosomes, one chromosome with heterochromatic blocks (h+h-) or chromosomes that lacked heterochromatic blocks (h-h-) (Fig. 2). Silver nitrate staining revealed the presence of multiple nucleolus organizer regions (NORs) located in the terminal portion of the short arm of two pairs of subtelocentric chromosomes (pairs 13 and 20) and in the terminal position of the long arm of an acrocentric chromosome pair (chromosome 34) (Fig. 1, in Box). In addition, a size heteromorphism was found in the Ag-NORs sites of chromosome pair 13 in all individuals analyzed. FISH with the 18S rDNA probe confirmed the silver nitrate staining result for pair 13, but only one of the homologues of the pairs 20 and 34 was marked (Fig. 3a). FISH revealed that the 5S rDNA sites were located in the interstitial portion of the short arm of a metacentric chromosome (pair 2) (Fig. 3b).
Karyotypes of female Hypostomus iheringii arranged from Giemsa-stained (a) and C-banded chromosomes (b). The chromosome pairs carrying Ag-NORs are boxed. Bar = 5 µm.
Accentuated heterochromatic polymorphisms on chromosome pairs 1 and 5 of the Hypostomus iheringii.
Karyotype of female Hypostomus iheringii submitted to FISH using 18S rDNA probe (a) and submitted to FISH using 5S rDNA probe (b). Bars = 5 µm.
Cytogenetic studies in Loricariidae reveal a remarkable diversity of chromosomal numbers, formulae and markers. Despite this extensive variation, karyotype analyses of the species in this family have allowed well-defined evolutionary trends and putative group relationships to be inferred (
The increase in the number of st/a chromosomes was postulated to be directly proportional to 2n while the number of m/sm chromosomes is inversely proportional to 2n. This hypothesis would suggest that centric fissions have played a key role in karyotype evolution of this group (
Polymorphisms of heterochromatic blocks with maintenance of heteromorphic states are relatively common among Teleost fishes and are correlated to population differentiation and speciation (
In Hypostomus iheringii, the polymorphism of the heterochromatic regions in chromosome pairs is not correlated to size variations in these euchromatic chromosomes (Fig. 2). Therefore, the process of heterochromatinization (inactivation by conversion of euchromatin into heterochromatin) might act on the long arms of chromosomes 1 and 5. However, the occurrence of additional amplification in these heterochromatic chromosomal regions cannot be ruled out completely.
Physical mapping of the 45S rDNA multigene family revealed a lack of staining of one homologue of chromosome pairs 20 and 34 (Fig. 3), probably because of unequal crossing over between homologues of these pairs resulting in sites of different sizes (
Although the literature on the physical mapping of 5S rDNA in Hypostomus is not abundant, variations within the group for this marker have been reported. This study identified only one chromosome pair carrying these sites, pair 2 (Fig. 3); however, eight chromosomes bearing such sequences were identified for Hypostomus affinis (
In summary, Hypostomus iheringii displays evolutionary trends that are characteristic of the genus Hypostomus, such as the high number of subtelocentric and acrocentric chromosomes assigned to the species of this genus with high chromosome numbers. However, the distribution and diversification of heterochromatin suggests new evolutionary trends. All chromosomal markers indicate that the karyotype of Hypostomus iheringii is highly differentiated and that heterochromatinization of chromosomal segments may contribute to karyotypic differentiation found in this Hypostomus iheringii population.
The authors are grateful to Prof. Dr. Heraldo A. Britski for the identification of the specimens, to the Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (IBAMA/MMA) for the authorization to collect the material; to Pedro L. Gallo e Luiz H. da Silva for collecting effort and to Prof. Dr. Luiz A. C. Bertollo for considerations. This study was financed by FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), Fundação Araucária (Fundação Araucária de Apoio ao Desenvolvimento Científico e Tecnológico do Estado do Paraná) and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico).