(C) 2012 Danon Clemes Cardoso. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
In this paper we present, for the first time, a detailed karyotype characterization of a species of the genus Azteca (Dolichoderinae, Formicidae). Cerebral ganglia from Azteca trigona Emery, 1893 were excised and submitted to colchicine hypotonic solution and chromosomal preparations were analyzed through conventional staining with Giemsa, C-banding, silver nitrate staining (AgNO3) and sequential base-specific fluorochromes. The analysis shows that Azteca trigona has a diploid number of 28 chromosomes. The karyotype consists of five metacentric pairs, seven acrocentric pairs and two pseudo-acrocentric pairs, which represents a karyotype formula 2K= 10M + 14A + 4AM and a diploid number of the arms 2AN = 38. The analysis of heterochromatin distribution revealed a positive block on distal region of the short arm of fourth metacentric pair, which was coincident with Ag-NOR band and CMA3 fluorochrome staining, meaning that rDNA sequences are interspaced by GC-rich base pairs sequences. The C-banding also marked short arms of other chromosomes, indicating centric fissions followed by heterochromatin growth. The karyotype analysis of Azteca trigona allowed the identification of cytogenetic markers that will be helpful in a difficult taxonomic group as Azteca and discussion about evolutionary aspects of the genome organization.
karyotype, chromosome number, chromosome banding, ants, Azteca trigona
The subfamily Dolichoderinae presents a great diversity of species throughout the world. Species are distributed in different biogeographic regions, from the Palearctic, Nearctic, Afrotropical region and Malaysia, to the Middle East, Australian and Neotropical regions (
The genus Azteca is strictly Neotropical and very diverse, including around 130 species. They are essentially arboreal and many species have mutualistic associations with particular plant species, where the genus Cecropia presents the most conspicuous association (
Cytogenetic characterization offers some of the most reliable taxonomic criteria for some groups of organisms and recently, the application of cytogenetic studies focused on understanding the distribution pattern and evolution of species seems very promising (
Even with the immense diversity of species of the genus Azteca, including approximately 130 described species, no cytogenetic study is encountered for this genus. Thus, to contribute to the increased cytogenetic knowledge of Formicidae and further understanding of karyotype evolution, the present study aimed to characterize the karyotype of Azteca trigona Emery, 1893, whereas karyotypes of the genera Anillidris Santschi, 1936 and Liometopum Mayr, 1861 remain totally unknown.
Material and methodsThirty specimens from two colonies of Azteca trigona collected in Ponte Nova (20°25'S, 42°54'W) and Viçosa (20°45'S, 42°52'W), MG, Brazil were analyzed. The colonies were collected in the field and transferred to a plastic container and maintained in a BOD (Biochemical Oxygen Demand) incubator at 25°C following the protocol described by
Cytogenetic analysis was performed using cerebral ganglia of the larvae selected. Metaphase chromosomes were obtained according to the methodology proposed by
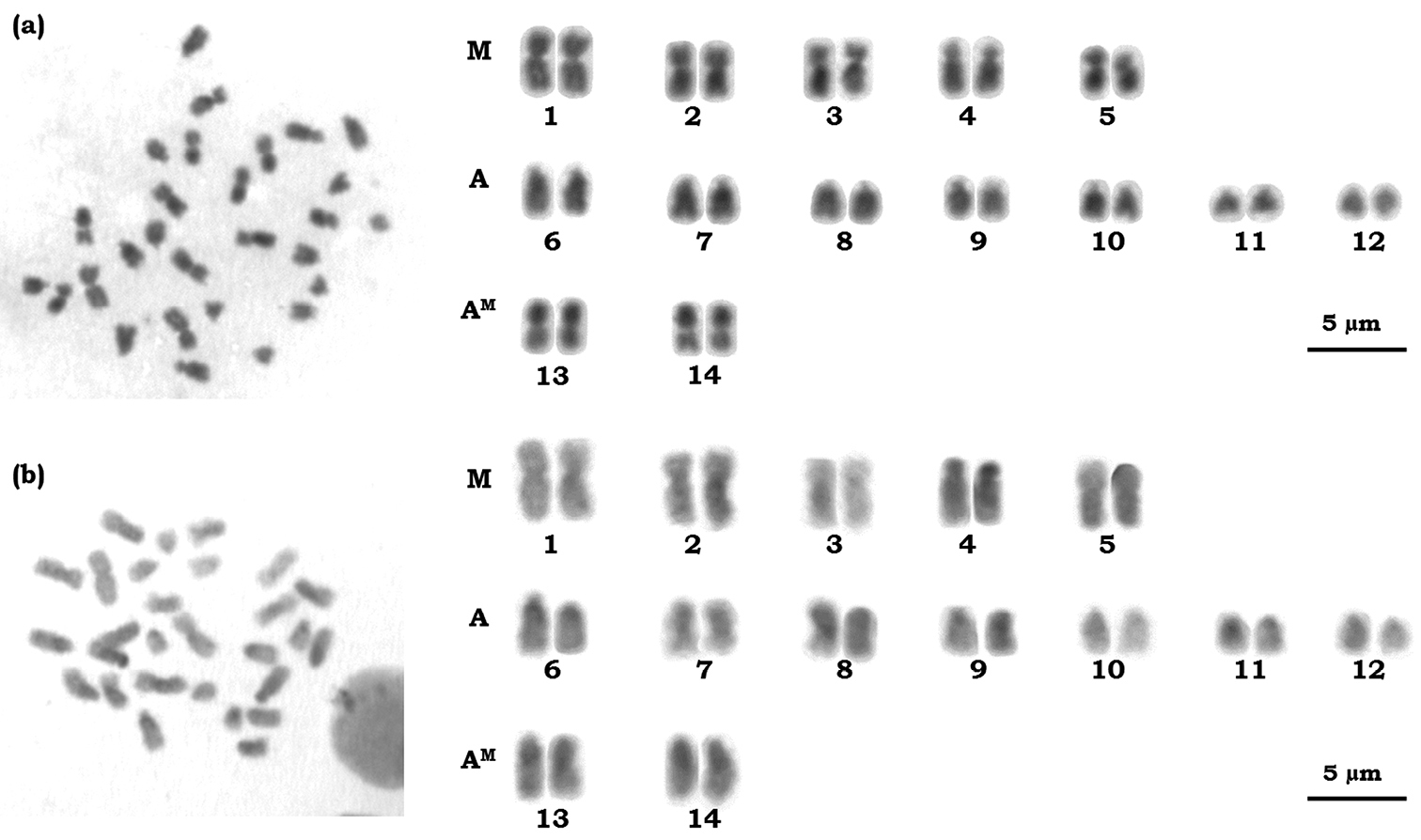
The chromosome number observed for Azteca trigona was 2n = 28 (Figure 1). The karyotype of this species consists of five metacentric pairs (M), seven acrocentric pairs and two pseudo-acrocentric pairs (AM) according to the terminology proposed by
Karyotype of female workers of Azteca trigona 2n = 28 sorted according to the classification proposed by
Of the subfamilies of Formicidae, Dolichoderinae is the fourth subfamily with the major number of taxa studied. According to
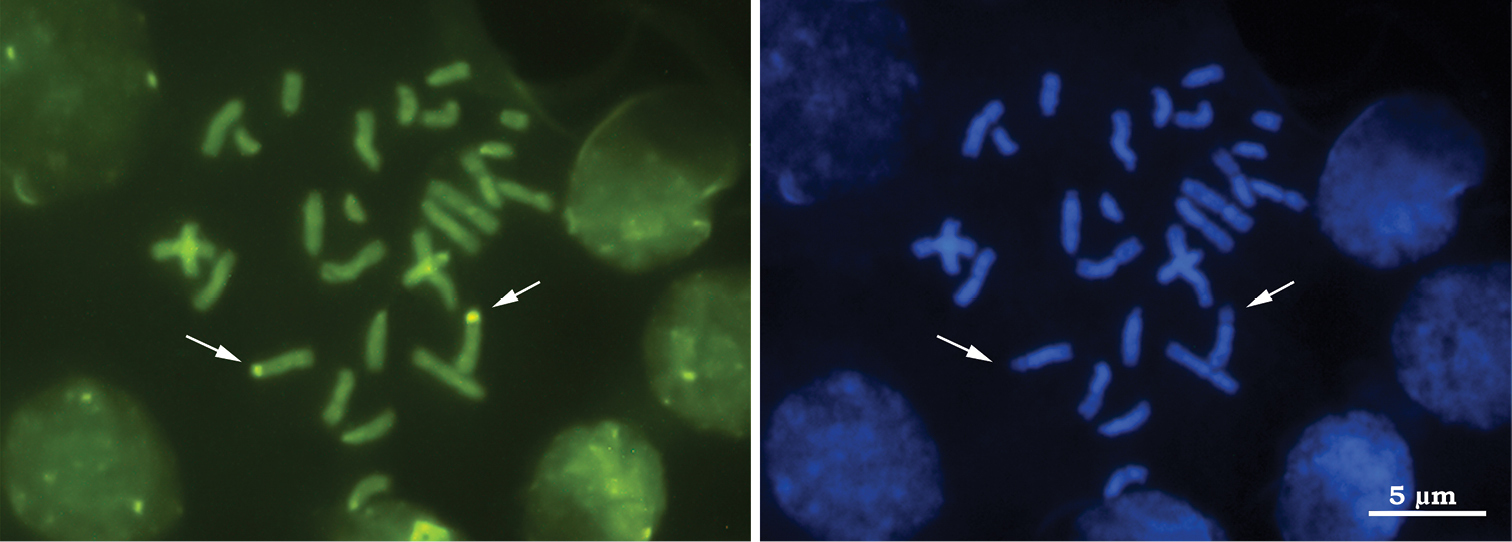
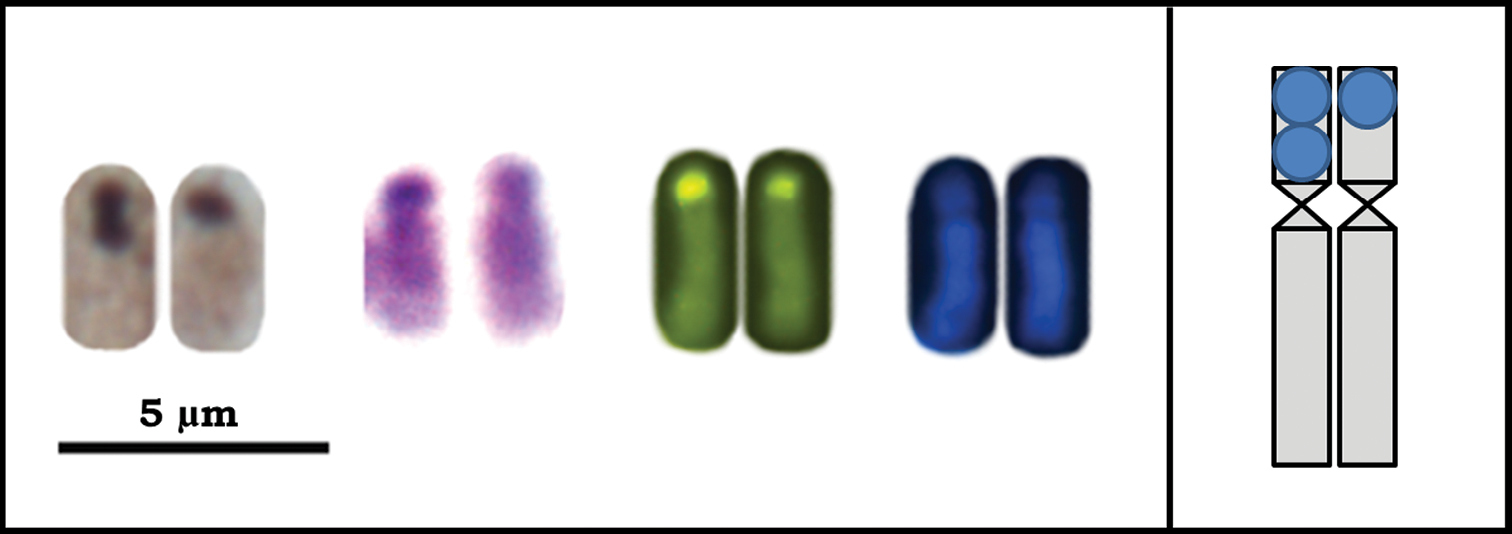
Results of the banding techniques indicate positive C-, Ag-NOR and CMA3-bands, and negative DAPI-bands on the short arm of the fourth pair of metacentric chromosomes (Figs 1–3), indicating that this region is rich in heterochromatin and should correspond to the nucleolus organizer region (NOR) (Fig. 3). This chromosome pair was CMA3-positive and DAPI-negative, indicating that the marked regions are rich in GC bases and devoid of AT bases (Fig. 2). Several authors have reported that CG-positive and AT-negative (i.e. auto-complementary) regions are related to nucleolus organizer regions (NORs). This relationship has been reported in grasshoppers (
Diploid metaphase of Azteca trigona submitted to sequential staining with fluorochromes CMA3/DA/DAPI a Staining with CMA3, arrows indicate the fourth chromosome pair and the GC+ regions b Staining with DAPI, arrows indicate the same chromosome pair with the negative AT-rich regions.
Furthermore, the silver nitrate staining revealed that the fourth metacentric pair also had an Ag-positive block. This finding corroborated that this pair carried NORs, which were heteromorphic between the homologues (Fig. 3). Heteromorphism in NOR size is frequent in a large number of organisms and can be explained by tandem duplications of the ribosomal genes (
The results presented here are, to our knowledge, the first cytogenetic data of a species of the genus Azteca, and the second known for a species of the subfamily Dolichoderinae in the Neotropics. Previously, cytogenetic data on the species Dorymyrmex pyramicus (Roger, 1863) (2n = 18) were presented on the base of only five workers collected in Uruguay (Gõni et al. 1983). According to some authors, cytogenetic data on Neotropical ant species are scarce given the immense biodiversity of this region (
In particular, the genus Azteca presents a great challenge to taxonomists since identification is practically impossible at the species level in absence of the queen (
Fourth pair of chromosomes of Azteca trigona submitted to different banding techniques: Ag-NOR, C-banding and sequential CMA3/DA/DAPI fluorochrome staining. The inserted scheme indicates that for all techniques the homologues are heteromorphic for the banding pattern.
The authors are grateful for the critical reading of two anonymous reviewers and Evan Vissar for language reviewing. We also thank Ricardo R. C. Solar for helping us with the ant sampling. This research was supported by Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). We also thank Rodrigo Feitosa and Jacques H.C. Delabie for taxonomic identification.