(C) 2012 Amina Kharrat-Souissi. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
The Buffelgrass (Cenchrus ciliaris L., Poaceae) is one of the most important pasturage grasses due to its high productivity and good forage qualities. This species possess a high adaptability to bioclimatic constraints of arid zones and may be used for the restoration of degraded arid ecosystems. Tunisian populations present three ploidy levels (4x, 5x and 6x) with a basic chromosome number x=9. This study reported for the first time the distribution of the ribosomal genes (rRNA) for pentaploid and hexaploid cytotypes of Cenchrus ciliaris. Molecular cytogenetic study using double fluorescence in situ hybridization has shown that the two rDNA families, 5S and 18S-5.8S-26S (18S), displayed intraspecific variation in number of loci among different ploidy levels. Each ploidy level was characterized by specific number of both 5S and 18S rDNA loci (two loci in tetraploid, five in pentaploid and six in hexaploid level). For three studied cytotypes (4x, 5x and 6x) all 5S rDNA loci were localized on the subcentromeric region of chromosomes, while 18S loci were situated on the telomeric region of short chromosome arms. Data of the FISH experiments show proportional increase of ribosomal loci number during polyploidization processes.
Buffelgrass, Cenchrus ciliaris, fluorescence in situ hybridization , fluorochrome banding, polyploidy, rDNA organization
In the south of Tunisia, the ecosystems are characterized by a high level of anthropogenic disturbance and have been characterized by several factors such as climatic variations and overgrazing (
Most flowering plants are polyploids, since polyploidization is a ubiquitous event in plant evolution (
The cytogenetic information provided by combination of chromosome banding and fluorescence in situ hybridization (FISH) can be useful for comparing the populations of the same species (
The objective of the current study was to elucidate the possible changes in number and location of rDNA sites through different ploidy levels of Cenchrus ciliaris by physical mapping of 5S and 18S rRNA genes.
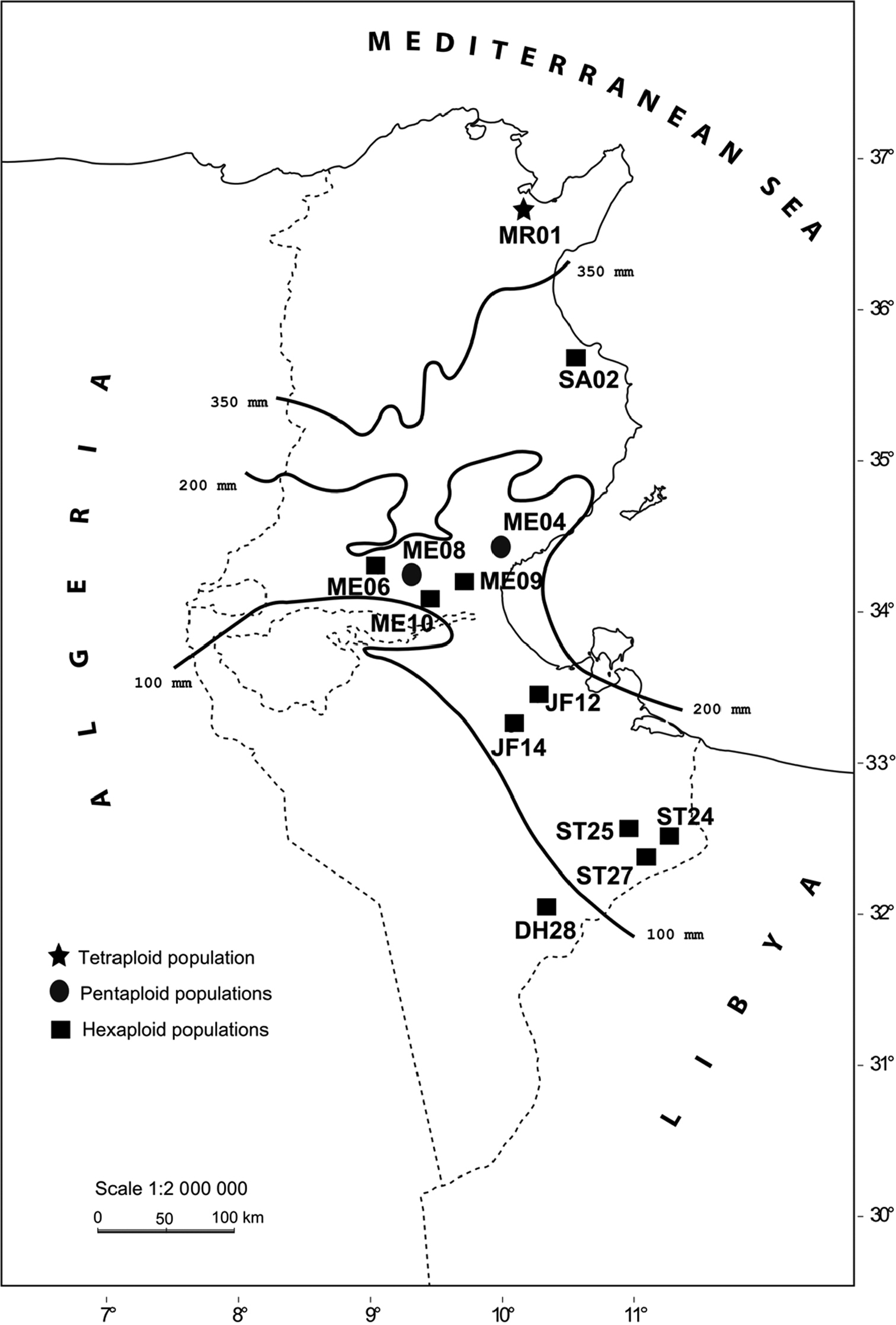
Material and methods Plant material and chromosome preparationThe geographical origins of Cenchrus ciliaris samples collected in natural populations in Tunisia are given in Table 1 and Fig. 1. The vouchers were deposited at the herbarium of the Laboratory of Plants Diversity and Ecosystems in Arid Areas, Department of Biology, University of Sfax. The seedlings were germinated on moist filter papers in Petri dishes at 28°C. After three days, the root tip meristems were removed from germinated seedlings and treated with 2 mM 8-hydroxyquinoline solution for 3 h at 16°C. Subsequently, the material was fixed in freshly prepared ethanol: acetic acid (3:1, v/v) solution.
Geographical origin, genome size, ploidy level, number of 5S and 18S rDNA loci in Tunisian populations of Cenchrus ciliaris. † Data from
| Localities | Population Code | Latitude | Longitude | 2C DNA in pg† | 2n† | Ploidy level (x)† | Number of 5S rDNA signals | Number of 18S rDNA signals |
| South of Tunisia city | MR01 | 36°73'N, 10°24'E | 3.03 ±0.03SD | 36 | 4 | 4 | 4 | |
| East of Teboulta | SA02 | 35°56'N, 11°06'E | 4.56±0.01 | 54 | 6 | 6 | 6 | |
| Meknassi Pist | ME04 | 34°32'N, 10°06'E | 3.74±0.09 | 45 | 5 | 5 | 5 | |
| Haddej Pist I | ME06 | 34°26'N, 09°12'E | 4.34±0.06 | 54 | 6 | 6 | 6 | |
| Haddej pist II | ME08 | 34°24'N, 09°29'E | 3.63±0.03 | 45 | 5 | 5 | 5 | |
| Gabès | ME09 | 34°10'N, 09°59'E | 4.55±0.03 | 54 | 6 | 6 | 6 | |
| El Hamma - Menzel Habib | ME10 | 34°02'N, 09°44'E | 4.46±0.09 | 54 | 6 | 6 | 6 | |
| Gabès- 45 Km – Medenin | JF12 | 33°37'N, 10°28'E | 4.57±0.05 | 54 | 6 | 6 | 6 | |
| Metameur-18Km- Toujane | JF14 | 33°24'N, 10°16'E | 4.49±0.11 | 54 | 6 | 6 | 6 | |
| IRA of Ben Guerdane-35 km-sidi Mahdi | ST24 | 32°49'N, 11°20'E | 4.47±0.04 | 54 | 6 | 6 | 6 | |
| National park of Sidi Toui (Est) | ST25 | 32°43'N, 11°14'E | 4.48±0.04 | 54 | 6 | 6 | 6 | |
| IRA of Ben Guerdane - 50 km - Sidi Mahdi | ST26 | 32°42'N, 11°18'E | 4.30±0.08 | 54 | 6 | 6 | 6 | |
| Remada-Dhibat (oued el Anguar) | DH28 | 32°08'N, 10°32'E | 4.34±0.04 | 54 | 6 | 6 | 6 | |
Geographical origin of 13 populations of Cenchrus ciliaris in Tunisia.
A slightly modified air drying technique (
GC-rich heterochromatin staining with chromomycin A3 (CMA3, Sigma Aldrich Co., Steinheim, Germany) was performed following
The FISH experiment was carried out with two different specific probes of ribosomal DNA (rDNA) simultaneously according to the protocol of
At least five metaphasic chromosome plates were used for karyometrical analysis and construction of idiogram. Chromosomes were classified according to their size and shape related to the centromere position. Terminology used for centromere position follows that of
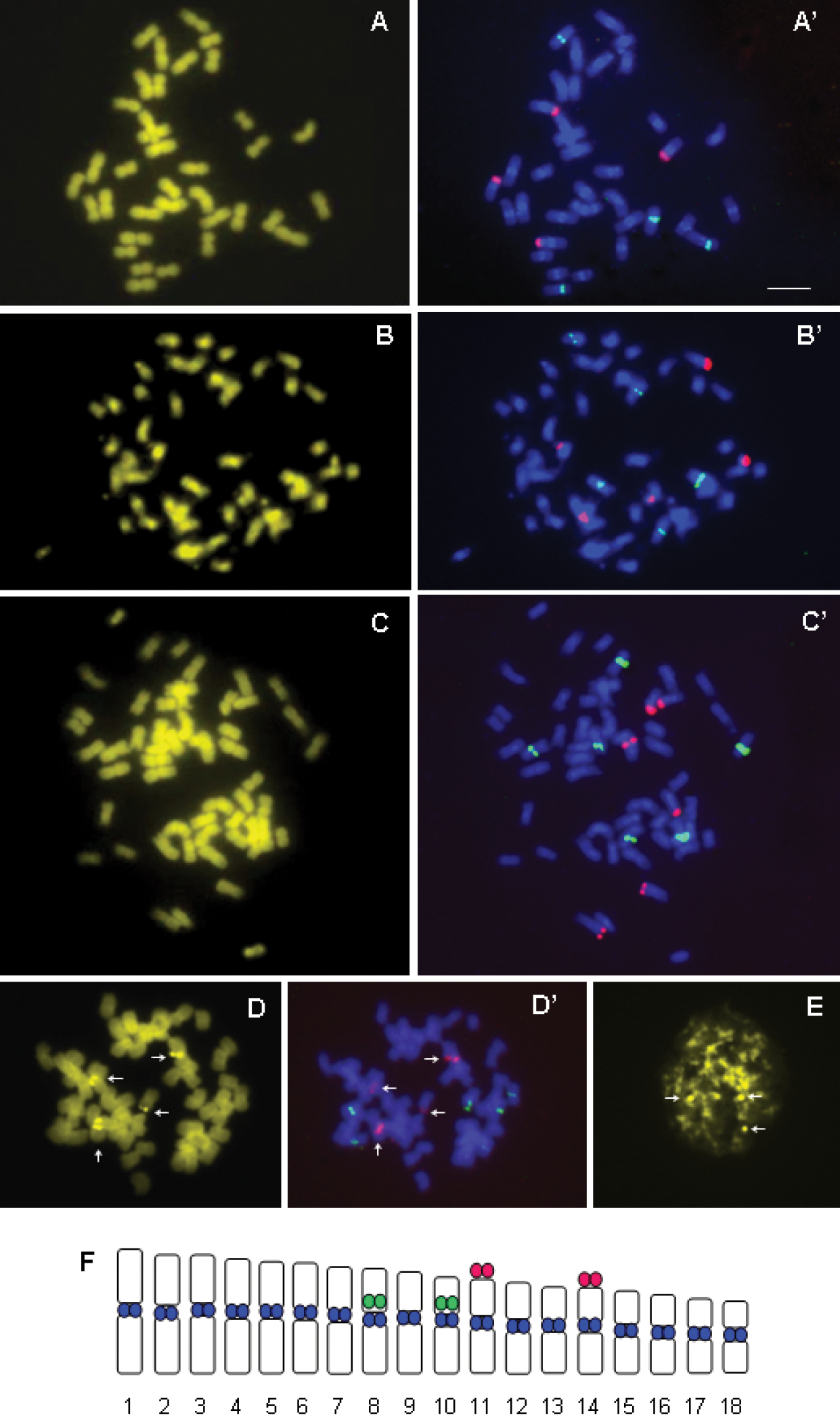
The chromosomes of Cenchrus ciliaris stained with CMA3generally showed the pale fluorescent bands of GC rich DNA detected with difficultyafter 90 min of staining, and not easily visible on microphotographs (Fig. 2A–B–C). These GC richbands were occasionally well visible only in hexaploids, where the maximum of three to four signals colocalized with 18S rDNA loci were observed (Fig. 2D). The chromomycin A3 positive signals were also observed in the interphase nuclei (Fig. 2E) which demonstrated the heterochromatin nature of this GC rich DNA. The clear centromeric DAPI+ bands, corresponding to constitutive heterochromatin, were observed after FISH experiments, but only for tetraploid population (Fig. 2A’–F).
A–F Chromomycin banding and fluorescence in situ hybridization of 5S rDNA (green) and 18S rDNA (red) probes respectively on the same chromosome plate: tetraploid individuals (A, A’); pentaploid individuals (B, B’) and hexaploid individuals (C, C’); CMA+ signals (D) correspond to 18S rDNA loci (D’) in hexaploid individuals; CMA+ signals in interphase nuclei (E); Idiogram of tetraploid individuals, with location of 5S (green), 18S (red) and DAPI (blue) signals (F). Bar = 10µm.
The results of 5S and 18S ribosomal genes mapping in Cenchrus ciliaris showed that tetraploid population exhibited four signals for both rRNA gene families (Fig. 2A’). In two studied pentaploid populations from different geographical origins (Table 1), five signals of both 5S and 18S rDNA were observed (Fig. 2B’), while six signals were detected in hexaploid populations (Fig. 2C’). It was observed that number of 5S and 18S rDNA loci increased, as expected, with ploidy level (Fig. 2): tetraploid individuals possessed four, pentaploids five, and hexaploids six loci. The 18S rDNA loci had terminal, while 5S rDNA presented pericentromeric localization (Fig. 2). The signals of 5S and 18S rDNA slightly vary in size and intensity, which was probably related to variation in number of copies. The results of CMA staining and FISH experiment on the same metaphase plate show that the GC rich regions, when they are detected as strong bands, correspond to the FISH signals of 18S rDNA probe (Fig. 2D–D’).
Chromosome identification and construction of idiogramBecause many of the chromosomes are similar in size and morphology, chromosome identification for karyotype analysis is very difficult for Cenchrus ciliaris. Thus, in present study we constructed the idiogram (Fig. 2F) for tetraploids based on conventional morphometry (Table 2) and determined the number and position of rRNA gene loci. The average chromosome length varied from 1.82 to 3.43 µm. The value of R (ratio between the longest and shortest chromosome pair, according to
Morphometric data concerning the karyotype of tetraploid Cenchrus ciliaris individuals. R = 0.9; AsI % = 47.89; s, short arm; l, long arm; c, total chromosome length; l/s, ratio of long and short arms; i, centromeric index = 100 x s/(l+s); m, metacentric chromosome type (according to
| Chromosome pair | s (µm) | l (µm) | c (µm) | (l/s) | i | Chromosome type |
| 1 | 1.64 (0.15) SD | 1.79 (0.12) | 3.43 | 1.09 | 47.92 | m |
| 2 | 1.54 (0.18) | 1.72 (0.12) | 3.27 | 1.12 | 47.27 | m |
| 3 | 1.46 (0.20) | 1.77 (0.11) | 3.23 | 1.21 | 45.30 | m |
| 4 | 1.48 (0.19) | 1.65 (0.14) | 3.13 | 1.11 | 47.29 | m |
| 5 | 1.42 (0.17) | 1.64 (0.13) | 3.06 | 1.16 | 46.36 | m |
| 6 | 1.42 (0.17) | 1.62 (0.12) | 3.04 | 1.14 | 46.76 | m |
| 7 | 1.34 (0.17) | 1.60 (0.13) | 2.94 | 1.19 | 45.59 | m |
| 8 | 1.34 (0.18) | 1.50 (0.12) | 2.84 | 1.12 | 47.17 | m |
| 9 | 1.21 (0.17) | 1.54 (0.14) | 2.75 | 1.26 | 44.16 | m |
| 10 | 1.17 (0.16) | 1.46 (0.13) | 2.63 | 1.24 | 44.56 | m |
| 11 | 1.13 (0.18) | 1.38 (0.09) | 2.51 | 1.23 | 44.84 | m |
| 12 | 1.13 (0.11) | 1.31 (0.09) | 2.44 | 1.17 | 46.15 | m |
| 13 | 1.05 (0.12) | 1.27 (0.10) | 2.32 | 1.20 | 45.38 | m |
| 14 | 1.04 (0.11) | 1.26 (0.09) | 2.29 | 1.22 | 45.14 | m |
| 15 | 1.01 (0.13) | 1.15 (0.13) | 2.16 | 1.14 | 46.69 | m |
| 16 | 0.98 (0.13) | 1.09 (0.12) | 2.07 | 1.11 | 47.41 | m |
| 17 | 0.91 (0.15) | 1.05 (0.11) | 1.96 | 1.16 | 46.36 | m |
| 18 | 0.84 (0.10) | 0.98 (0.12) | 1.82 | 1.17 | 46.08 | m |
The GC-rich DNA regions detected in hexaploids are distributed in telomeric regions of chromosomes and corresponded to the 18S rDNA loci. This colocalization of GC rich heterochromatin and rDNA has already been reported for numerous plants and animal species (
It was obvious to notice that the number of 5S and 18S rDNA sites corresponded to the ploidy level. In tetraploid individuals it was four, in pentaploids five and in hexaploids six signals. Similar phenomenon occurred in polyploids of some other genera, such as Saccharum Linnaeus, 1753 (
Tetraploid individuals of Cenchrus ciliaris show four signals for both 5S and 18S, the same result obtained by
A comparison of hybridization patterns between the two probes revealed identical results within each ploidy level of Cenchrus ciliaris. Thus the number of 18S and 5S rDNA loci in different individuals of each ploidy level was constant. This can be explained by the genome stability occurring in the three ploidy levels. This highly conserved nature of both 5S and 18S rDNA loci during polyploid evolution within Cenchrus ciliaris is not in accordance with observations made in hexaploid of Ipomea batata, indeed within this cytotype some varieties presented 18 signals of 18S and other 12 signals (
In analyzed individuals of Cenchrus ciliaris all 5S rDNA loci were localized on paracentromeric region of chromosome pairs 8 and 10, while 18S loci were situated on telomeric region of short chromosome arm of pairs 11 and 14.
The size and the intensity of both hybridization signals slightly varied among investigated individuals. This can be explained by different copy number of repeats among rDNA sites which has been also detected in several other plant species (
The distribution of investigated populations of Cenchrus ciliaris in Tunisia follows a north-south bioclimatic gradient, where ploidy level and genome size are increasing with aridity (
In the present study the number of signals of 5S and 18S rDNA loci in pentaploids was intermediate between tetraploids and hexaploids. This result seems to indicate that pentaploid individuals might have derived from hybridization events between tetraploids and hexaploids. Although the apomictic mode of reproduction known as apospory displayed by most Cenchrus ciliaris genotypes, rare sexual individuals have been identified (
Our data of the FISH experiments show proportional increase of ribosomal loci number during polyploidization processes. However ploidy level increases with aridity (from tetraploid to hexaploid) and give a cytogenetic basis to the considerable differentiation noted between north (humid area) and south (arid area) Tunisian populations of Cenchrus ciliaris.
The authors are grateful to O. Robin for assistance in cytogenetic and to A. Vucicevic for linguistic help. They are also indebted to the Ministry of Education and Science of Tunisia, CNRS (Centre National de la Recherche Scientifique) and Comité Mixte de Coopération Universitaire France-Tunisie (CMCU, project n° 09G 0901) for their financial support.