(C) 2012 Ana Heloisa de Carvalho. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
In this work we present a new karyotype for Rhipidomys Tschudi, 1845 (Cricetidae, Rodentia) from Brazil. Our chromosome analyses included GTG- and CBG-banding patterns, the localization of the nucleolus organizer regions after silver staining (Ag-NORs) and fluorescence in situ hybridization (FISH) with a telomere probe. The new karyotype is composed of 44 chromosomes and has a fundamental number (number of autosomal arms) of 48. Most Rhipidomys species already karyotyped presented similar complements with 2n=44, but their fundamental numbers varied from FN=46 to 80, a variation that has been mainly attributed to pericentric inversions. The comparison of this new karyotype to those of other Rhipidomys already reported allowed us to conclude that it is a distinctive chromosome complement, which can be of great use as a tool for the very complicated taxonomic identification in this genus.
Rhipidomys, chromosome banding, FISH
The Neotropical rodent Rhipidomys Tschudi, 1845 (family Cricetidae) is an arboreal genus belonging to the largely diverse subfamily Sigmodontinae, whose phylogenetic relationships are difficult to resolve, resulting in taxonomic uncertainties at every level, from species to tribes (
Rhipidomys is widely distributed and has been reported from Panama to southeastern Brazil and northern Argentina. The distribution of many species remains uncertain and there are several reports of undescribed species (
Eleven species of Rhipidomys have already been karyotyped and, with the exception of Rhipidomys nitela (2n=48) and Rhipidomys prope nitela (2n=50), all presented karyotypes with 2n=44 chromosomes. In contrast with the conservation of diploid numbers, the karyotypes of Rhipidomys showed fundamental numbers ranging from FN=46 to 80, a variation mainly attributed to pericentric inversions. The available karyotypical data for Rhipidomys are summarized in Table 1. Most cytogenetic studies on this genus were performed with conventionally stained karyotypes and in less than half the GTG- CBG- or AgNOR-banding patterns were also included.
Table 1. Summary of the available chromosome data for Rhipidomys.1 As Rhipidomys sclateri, which was later considered a synonym of Rhipidomys leucodactylus (
| Group | Species | 2n/FN | Locality | Reference |
|---|---|---|---|---|
| Rhipidomys leucodactylus | Rhipidomys leucodactylus | 44/ 46 | Rio Juruá (AM) | |
| 44/ 48 | Rio Jamari (RO), Caldas Novas, Serra da Mesa (GO) |
|
||
| 44/ 481 | Cueva del Agua (Venezuela) | |||
| 44/ 52 | Serra da Mesa (GO), Caxiuanã (PA) | |||
| Rhipidomus sp. | 44/ 48 | Berilo (MG) | This work | |
| Rhipidomys latimanus | 44/ 48 | Peñas Blancas (Colômbia) | ||
| Rhipidomys macrurus | 44/ 48 | Águas Emendadas (DF), Chapada Diamantina (BA) |
|
|
| 44/ 49 | Granja do Ipê (DF) | |||
| Rhipidomys prope macrurus | 44/ 492 | Casa Grande (SP) | ||
| 44/ 502 | Monte Verde (ES) | |||
| 44/ 50 | Garrafão (RJ) | |||
| 44/ 51 | Mocambinho (MG) | |||
| Rhipidomys gardneri | 44/ 50 | Rio Juruá (AC) | ||
| Rhipidomys macconnelli | 44/ 50 | La Escalera (Venezuela) | ||
| Rhipidomys cf. mastacalis | 44/ 52 | Vila Rica (MT), Aripuanã (MT) | ||
| Rhipidomys itoan | 44/ 48, 49, 50 | SP and RJ | ||
| Rhipidomys mastacalis | Rhipidomys mastacalis | 44/ 74 | Lagoa Santa (MG), Unacau (BA), Casimiro de Abreu (RJ), Reserva Biológica Duas Bocas (ES) |
|
| 44/ 76 | Serra da Mesa (GO) | |||
| 44/ 80 | Serra da Mesa (GO) | |||
| 344/ high | Serra dos Cavalos (PE) | |||
| Hybrid | Rhipidomys with high FN x Rhipidomys with low FN | 44/ 61 | M. Chapéu (BA) | |
| Rhipidomys nitela | Rhipidomys nitela | 44/ 71 | San Ignacio, (Venezuela) | |
| 48/ 67 | La Trinité (French Guiana) | |||
| 48/ 68 | Surumurú (RR) | |||
| Rhipidomys prope nitela | 50/ 71, 72 | Manaus (AM) |
In this work, we present a new karyotype for Rhipidomys. Our analyses included GTG- and CBG-banding patterns, the silver staining location of the nucleolus organizer regions (Ag-NORs) and fluorescence in situ hybridization (FISH) with a telomere probe.
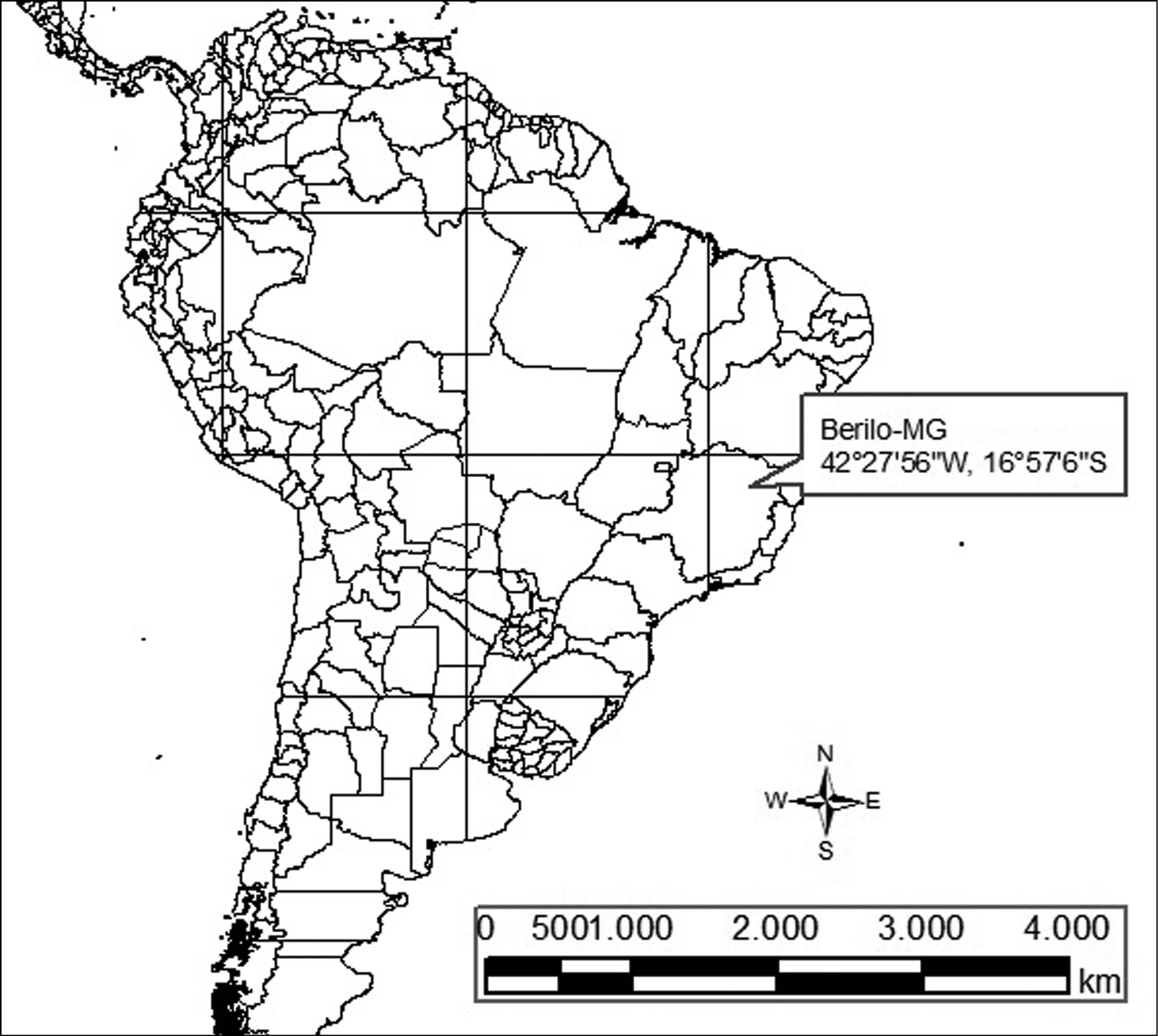
Material and methodsWe analyzed five specimens (two males and three females) of Rhipidomys sp. captured in 2004 in a dry land region in the margins of the Jequitinhonha river, in Berilo, state of Minas Gerais, Brazil (16°57'06"S, 42°27'56"W; Fig. 1) under the license 129/04-NUFAS/MG from the Instituto Brasileiro para o Meio Ambiente - IBAMA. The skins and skulls were deposited at the Museu de Ciências Naturais da Pontifícia Universidade Católica de Minas Gerais, in Belo Horizonte, Minas Gerais State, Brazil, under the numbers: MCNM 1643, 1644 (two males) and MCNM 1646, 1647, 1648 (three females).
Map showing the collection locality of the Rhipidomys sp. analyzed.
Chromosome preparations were obtained from bone marrow according to the technique described by
The chromosomes were arranged based on the karyotype described for specimens of Rhipidomys sp. by
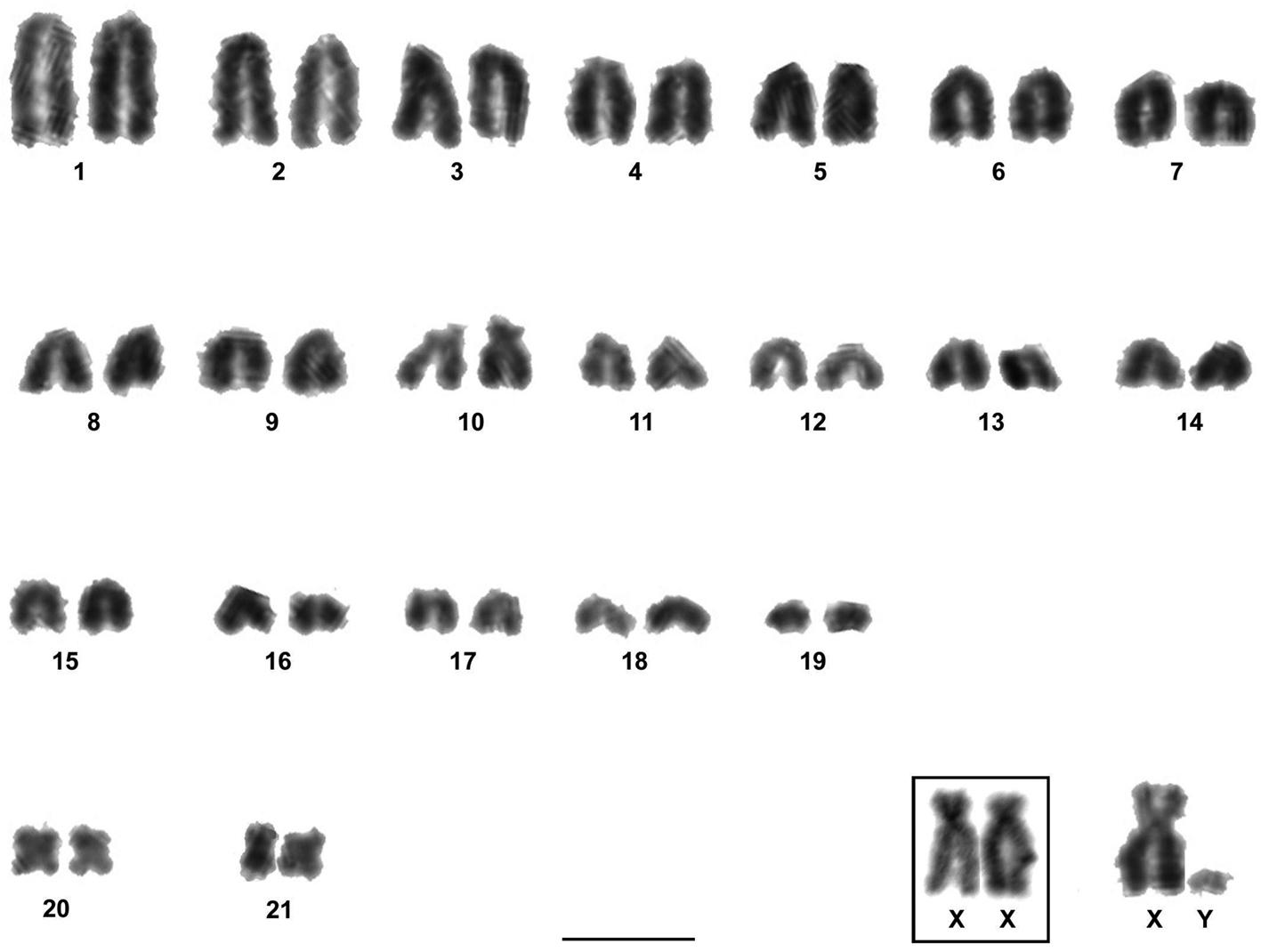
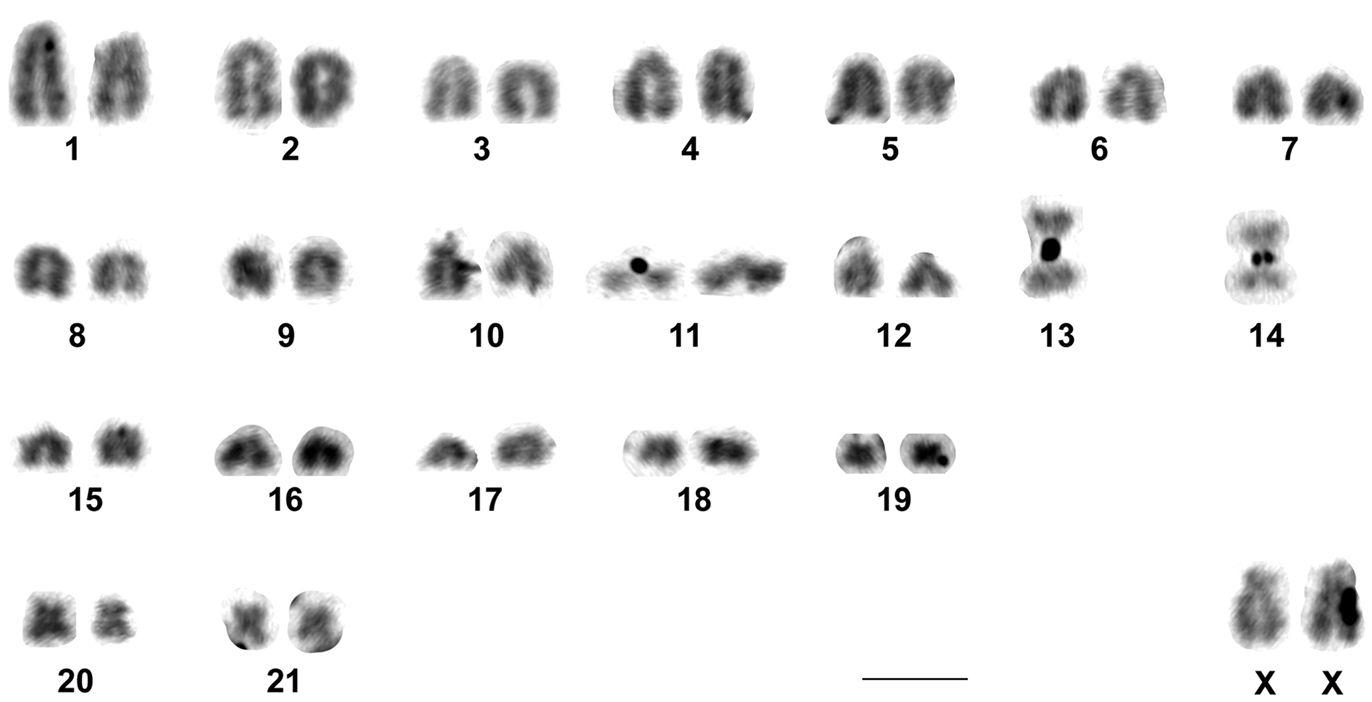
The two males and three females of Rhipidomys sp. analyzed presented a diploid number of 2n=44 chromosomes and a fundamental number FN=48. This karyotype was composed of 21 pairs of autosomes: 18 pairs of acrocentrics with gradual variation in size from large to small (pairs 1 to 9 and 11 to 19), one pair of medium subtelocentrics (pair 10), one pair of small metacentrics (pair 20) and one pair of small submetacentrics (pair 21). The X chromosome was a large submetacentric with polymorphism in the size of its short arms and the Y chromosome was a very small acrocentric. Autosomal pairs 1, 10, 19, 20 and 21, the X and the Y chromosomes were the only identifiable chromosomes after conventional Giemsa staining (Fig. 2).
Karyotype of Rhipidomys sp. male (2n=44, FN=48) after conventional Giemsa staining. In the inset, the sex chromosomes of a female. Note the variation in the size of the short arms of the X chromosomes. Bar = 10 µm.
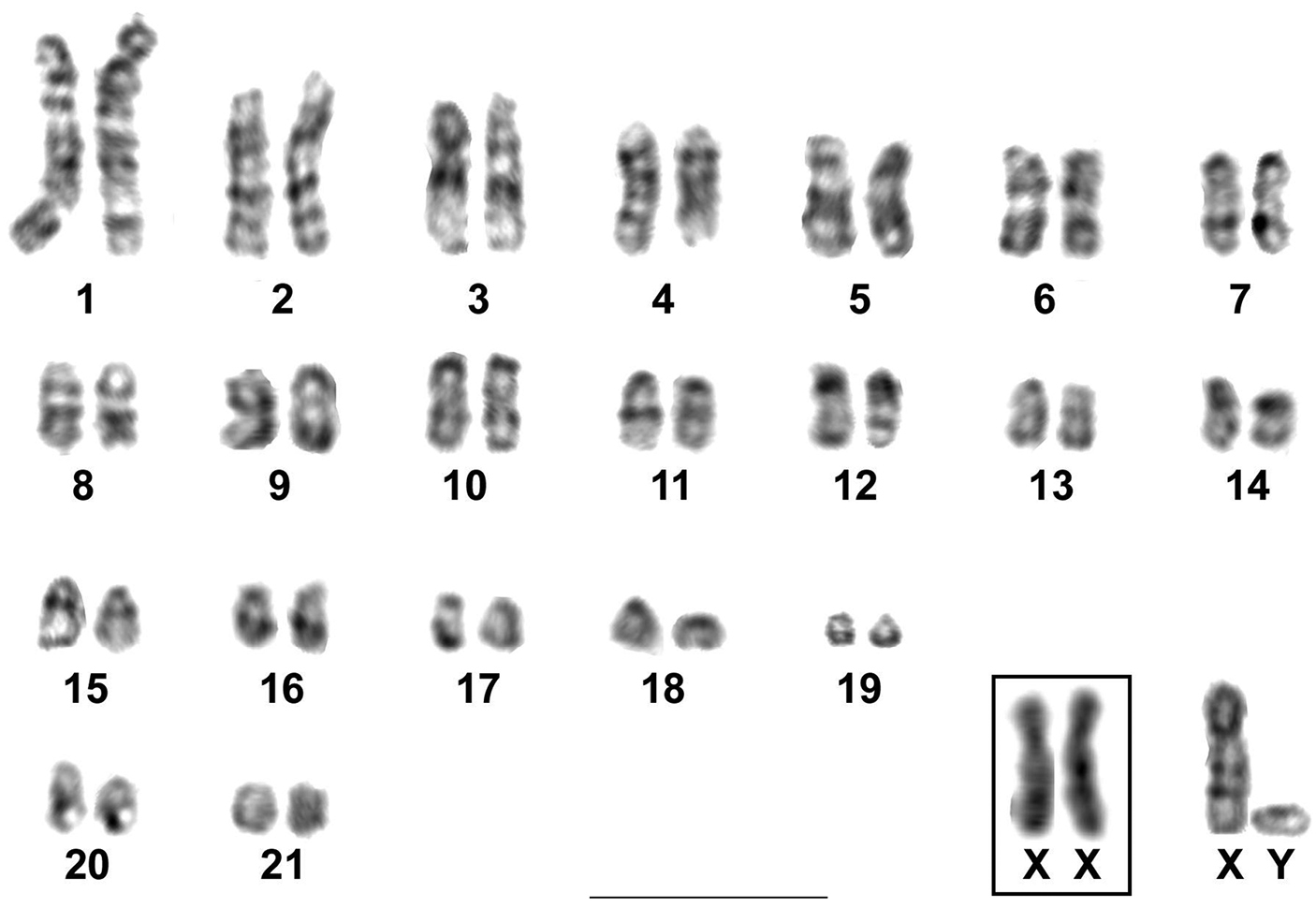
After GTG-banding all the autosomes and the sex chromosomes could be identified. The X chromosome presented the two typical mammalian dark GTG-bands in its long arm and no bands were observed on its short arms. The Y chromosome had an indistinct staining (Fig. 3).
GTG-banded karyotype of Rhipidomys sp. male (2n=44, FN=48). In the inset, the sex chromosomes of a female. Bar = 10 µm.
CBG-banding revealed the presence of constitutive heterochromatin in the pericentromeric region of most autosomal pairs. The short arm of the X chromosome was entirely heterochromatic with a stronger stained pericentromeric region and the Y chromosome displayed a small pericentromeric C-band (Fig. 4).
CBG-banding in a metaphase of Rhipidomys sp. female (2n=44, FN=48). Bar = 10 µm.
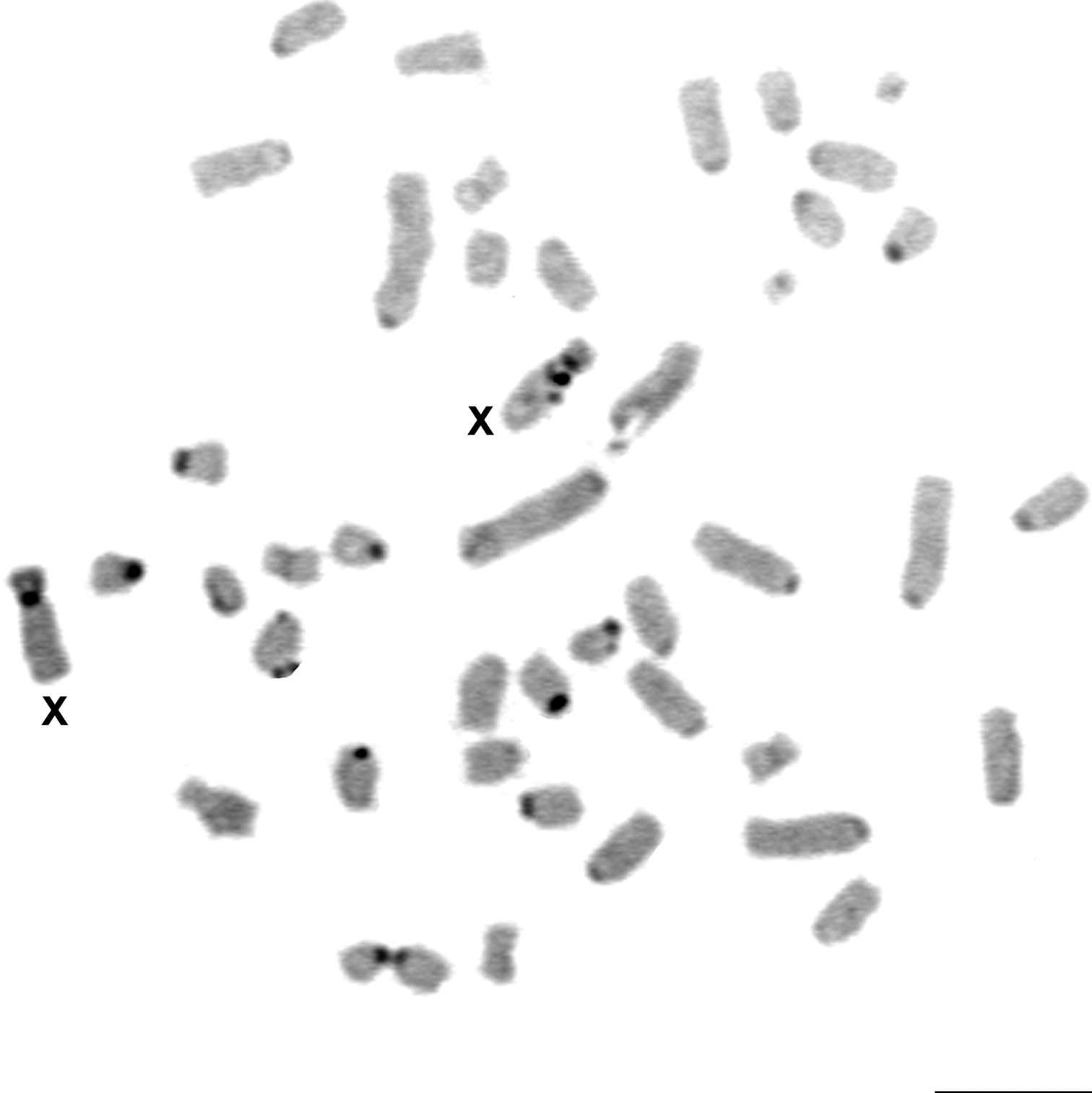
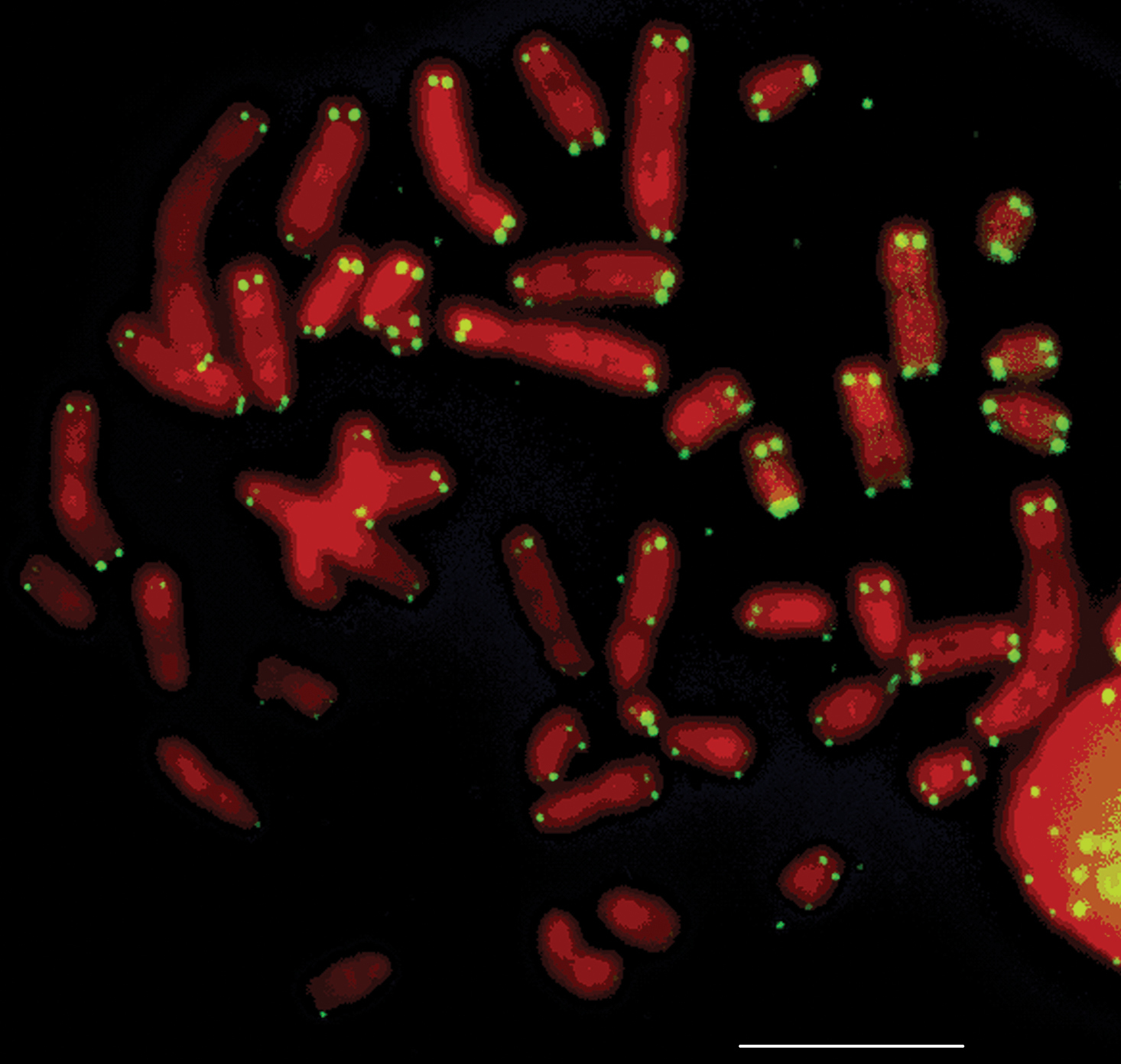
Silver staining revealed one to five nucleolus organizer regions (Ag-NORs) per cell. The Ag-NORs were located on the short arms of medium/small acrocentric autosomes. From the 151 analyzed cells, the majority (57) showed four Ag-NORs. Associations between NORs were frequent (Table 2, Fig. 5). FISH with the telomere sequences revealed signals only at the telomere regions of all chromosomes (Fig. 6).
Number of Ag-NORs per cell in Rhipidomys sp. (2n=44, FN=48).
| Number of chromosomes with Ag-NORs | |||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | Total | ||
| Number of cells | MCNM 1643 (Male) | 4 | 5 | 10 | 10 | 1 | 30 |
| MCNM 1644 (Male) | 2 | 6 | 6 | 13 | 3 | 30 | |
| MCNM 1646 (Female) | 5 | 4 | 10 | 7 | 4 | 30 | |
| MCNM 1647 (Female) | 1 | 3 | 16 | 11 | 0 | 31 | |
| MCNM 1648 (Female) | 1 | 0 | 12 | 16 | 1 | 29 | |
| Total | 13 | 18 | 54 | 57 | 9 | 151 | |
Silver staining of the nucleolus organizer regions (Ag-NORs) in the karyotype of Rhipidomys sp. female (2n=44, FN=48). Bar = 10 µm.
Metaphase of Rhipidomys sp. female (2n=44, FN=48) after FISH with a telomere probe. Bar = 10 µm.
Besides the karyotype of Rhipidomys sp. presented herein, four other species of Rhipidomys with the karyotype formula of 2n=44 and FN=48 have been described: Rhipidomys latimanus Tomes, 1860 from Colombia, for which no karyotype picture has been presented (
The karyotype of Rhipidomys sp. studied herein differed from that of Rhipidomys macrurus (2n=44, FN=48) from Goiás (
GTG-banding patterns have not been described for Rhipidomys leucodactylus, also with 2n=44 and FN=48. From the three biarmed autosomes found in the karyotype of this species, two are comparable in size to the medium acrocentric pair 15 and the third is the smallest autosome pair (
In Rhipidomys itoan with 2n=44 and FN=48 the smallest autosome pair was a submetacentric (
The absence of banding patterns descriptions limited the comparisons of the complement of R. sp. described in this work and those of Rhipidomys itoan and Rhipidomys leucodactylus to conventionally stained chromosomes.
The Rhipidomys species already recorded in Minas Gerais were Rhipidomys macrurus, which is probably distributed in the remaining Cerrado fragments of the state, Rhipidomys mastacalis, which was collected in the Atlantic Forest in eastern and southern Minas Gerais, and Rhipidomys tribei, known from only a few sites in the southeastern part of Minas Gerais (
Rhipidomys mastacalis is characterized by a high fundamental number (FN=74 through 80) (
FISH with telomere sequences has been previously performed in specimens of Rhipidomys nitela, Rhipidomys mastacalis and Rhipidomys leucodactylus (
The identification of Rhipidomys specimens from southeastern Brazil at the species-level has proven to be specially challenging, with Rhipidomys macrurus and Rhipidomys mastacalis being among the most taxonomically complicated taxa studied (
Chromosome analyses may be useful for the identification of species, especially in complicated taxonomic groups, as is the case of many rodent taxa. As stressed by
With the available data, it seems evident that a larger collection effort including a wider geographical range and complemented by cytogenetic and molecular studies will be needed in order to establish the phylogenetic relationships and phylogeography of Rhipidomys in Brazil. Nevertheless, as exemplified in this work, the use of chromosome data has already proven to be a useful tool in resolving taxonomic issues in this genus.
This work was supported by Fundação de Amparo à Pesquisa do Estado de Minas Gerais, Fapemig (processes CBB APQ-1495-3.12/07, CRA - APQ-00170-09 and APQ-00336-09) and by a grant of the Pró-Reitoria de Pesquisa, UFMG. AHC was a recipient of a graduate fellowship from CAPES, Brazil. The authors are indebted to the Laboratório de Citogenética, Hospital das Clínicas, UFMG, for the use of their laboratory and to Bárbara MA Costa, for the useful discussion of the manuscript.
References