(C) 2012 Santosh K. Sharma. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Understanding the genetic resources and diversity is very important for the breeding programs and improvement of several economically important orchids like Cymbidium. Karyomorphological studies have been carried out on seven Cymbidium species, Cymbidium aloifolium (Linnaeus, 1753), Cymbidium devonianum Paxton, 1843, Cymbidium elegans Lindley, 1828, Cymbidium iridioides D. Don, 1825, Cymbidium lowianum Rchb. f., 1877, Cymbidium tigrinum Parish ex Hook. f., 1864, and Cymbidium tracyanum L. Castle, 1890, most of them endangered/threatened in their natural habitat. As reported earlier, the somatic chromosome number (2n = 40) has been observed in all the seven species. Distinct inter-specific variation was recorded in the arm ratio of few homologous pairs in the complements. Symmetrical or almost symmetrical karyotypes were prevalent; however significant asymmetry was reported in Cymbidium iridioides and Cymbidium tracyanum. The significance of karyotypic variation in speciation of the genus Cymbidium has been discussed. This study provides useful chromosome landmarks and evidence about genome evolution, heteromorphic chromosomes based heterozygosity, basic chromosome number and ploidy level in the genus Cymbidium.
Orchidaceae, mitosis, karyotype, heteromorphism, symmetry
Cymbidium, or boat orchid, is a myriad orchid with evergreen foliage and arching sprays of delicately colored and waxy flowers, comprising of 52 evergreen species in the subtribe Cyrtopodiinae of tribe Cymbidieae (Orchidaceae). Cymbidiums are renowned for an abundance of morpho-types, with a seemingly unending array of strange and often impressive variations, and represent a highly advanced terminal line of floral evolution in the family. The genus is characterized by a broad geographical distribution encompassing tropical and subtropical Asia, South of Papua, New Guinea and Northern Australia, and exhibits a tremendous diversity in growth habits. It comprises several such representatives capable of occupying almost every conceivable ecological situation, apart from marine environments and habitats characterized by extreme cold throughout the year. Inter-generic compatibility is giving rise to hybrid groups, which are characterized by both greater size and hybrid vigor vis-à-vis their putative parental species. Therefore, characterization of genetic resources and diversity is a clue for framing meaningful breeding programs of economically important orchids like Cymbidium (
A number of workers from Asiatic regions especially China and Japan focused on cytogenetical aspects of several Cymbidium species: Cymbidium cyperifolium Lindly, 1833, Cymbidium faberi Rolfe, 1896, Cymbidium goeringii Rchb. f., 1852, Cymbidium kanran Makino, 1902, Cymbidium longibracteatum Y.S. Wu et S.C. Chen, 1966, Cymbidium qiubeiense K.M. Feng et Li, 1980and Cymbidium serratum Schlechter, 1919(
The karyomorphological details of Indian representatives of Cymbidium are still ambiguous, which make it difficult to correctly estimate ploidy levels vis-à-vis karyological evolution. In addition to our earlier efforts (
Cytological data on the Indian orchid flora are available for relatively few genera and most of them are restricted to chromosome counts only (Arora 1960,
The young plants belonging to seven species of Cymbidium were collected mainly from Arunachal Pradesh, Meghalaya and Sikkim provinces of Northeastern region of India. The plants were grown in greenhouses of North-Eastern Hill University, Shillong. For each species, a minimum of five individuals belonging to more than one population were studied. Details regarding collection of root tips, staining, chromosome complement preparation and their analysis are as described than described in
The seven Cymbidium species presently investigated show the diploid number of 2n = 40 chromosomes in root tip cells, which were clearly resolved into 20 pairs forming a series from the longest to shortest pair within the complements. The details of karyomorphological aspects including pair-wise arm ratio, karyotypic formula, number of sub-telocentric chromosome and/or heteromorphic pairs are illustrated in Tables 1–2 and Figs 1–14. One notable feature was the lack of distinct nucleolar chromosomes in any of the seven species investigated. Variation was recorded with respect to the number of metacentric and submetacentric chromosomes, presence or absence of heteromorphic pairs in the chromosome complements ofall the seven species of Cymbidium (Table 1). This study revealed that the plants belonging to Cymbidium tracyanum are peculiar in presenting metacentric and/or submetacentric chromosomes with one pair of distinct subtelocentric chromosomes in the complement. On the other hand, all the other cymbidiums are characterized by having exclusively submetacentric and/or metacentic chromosome pairs in karyotypes and are devoid of any subtelocentrics (Table 1).The chromosome morphology with regard to a particular pair in the karyotype has shown significant variation at inter-specific level (Table 2, Figs 8–14). For example, the third pair of Cymbidium lowianum and Cymbidium tracyanum is metacentric whereas all other cymbidiums have sub-metacentric chromosomes for this particular pair. The fifth pair in Cymbidium tracyanum, is found to be sub-telocentric whereas, in other cymbidiums it is either metacentric or sub-metacentric. Such observation can be extended even too other pairs (i.e. IV-VIII, XIII, XVII, and XX) as well. Except for these, the rest of the pairs were found to be exclusively sub-metacentric (Table 1). Chromosome pairs VI, IX, XIV, XV, XVI, XVII, XVIII and XIX are found to be heteromorphic in Cymbidium aloifoium, Cymbidium devonianum, Cymbidium elegans, Cymbidium lowianum, Cymbidium mastersii, Cymbidium tigrinum and Cymbidium tracyanum, respectively. The highest number of heteromorphic pairs i.e. three (XV, XVII, and XVIII) are recorded in Cymbidium elegans, (Fig. 11) followed by Cymbidium tracyanum (Fig. 14) which had two heteromorphic pairs (XVII and XIX). Alternatively, not a single pair of the chromosome was found to be heteromorphic in Cymbidium iridioides (Table 1 and Fig. 13).
Following the classification of
Karyomorphology and arm ratio in Cymbidium species.
| Taxa | 2n | r-index in different chromosomes | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII | XIII | XIV | XV | XVI | XVII | XVIII | XIX | XX | ||
| Cymbidium tigrinum | 40 | 1.21 L | 1.21 L | 1.21 L | 1.04 V | 1.04 V | 1.46 L | 1.2 L | 1.33 L | 2.54 L | 1.18 L | 1.84 L | 1.36 L | 1.29 L | 1.18 L | 1.45 L | 2.03 L | 1.43 L | 1.27 L | 1.48 L | 1.5 L |
| Cymbidium lowianum | 40 | 1.44 L | 1.12 L | 1.09 V | 1.23 L | 1.16 L | 1.14 L | 1.24 L | 1.6 L | 1.2 L | 1.29 L | 1.37 L | 1.14 L | 1.19 L | 1.57 L | 1.34 L | 1.12 L | 1.08 V | 1.38 L | 2.26 L | 1.06 V |
| Cymbidium devonianum | 40 | 1.12 L | 1.25 L | 1.20 L | 1.44 L | 1.35 L | 1.27 L | 1.06 V | 1.7 L | 1.32 L | 1.33 L | 1.34 L | 1.33 L | 1.7 L | 1.48 L | 1.25 L | 1.16 L | 1.62 L | 1.17 L | 1.55 L | 1.35 L |
| Cymbidium elegans | 40 | 1.24 L | 1.37 L | 1.1 L | 2.67 L | 1.21 L | 1.09 V | 1.25 L | 1.22 L | 1.46 L | 1.24 L | 1.25 L | 1.59 L | 1.09 V | 1.32 L | 1.54 L | 1.26 L | 1.43 L | 1.15 L | 1.33 L | 1.23 L |
| Cymbidium aloifolium | 40 | 1.63 L | 1.33 L | 2.46 L | 1.97 L | 1.36 L | 2.5 L | 1.06 V | 1.08 V | 1.9 L | 2.2 L | 1.67 L | 1.3 L | 1.08 V | 1.13 L | 1.22 L | 1.19 L | 1.16 L | 1.35 L | 1.72 L | 1.32 L |
| Cymbidium iridioides | 40 | 1.22 L | 1.29 L | 1.46 L | 1.44 L | 1.07 V | 1.11 L | 1.06 V | 1.52 L | 1.27 L | 2.27 L | 1.7 L | 1.02 L | 1.18 L | 1.31 L | 1.26 L | 1.28 L | 1.64 L | 1.15 L | 2.2 L | 1.25 L |
| Cymbidium tracyanum | 40 | 1.64 L | 1.2 L | 1.02 V | 1.08 V | 3.24 J | 1.05 V | 1.16 L | 1.61 L | 1.25 L | 1.8 L | 1.18 L | 1.06 V | 1.58 L | 1.67 L | 2.11 L | 2.3 L | 2.17 L | 1.47 L | 1.21 L | 1.23 L |
Underlined values are showing heteromorphic pairs of chromosomes.
Chromosome characteristics in various Cymbidium species.
| Taxa | Range SC-LC (μm) | Ratio LC/SC | p (μm) Mean (±SD) | q (μm) Mean (±SD) | CL (μm) Mean (±SD) | CI Mean (±SD) | CVCL | CVCI | AI ( |
Category of symmetry ( |
Karyotypic formula ( |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cymbidium tigrinum | 2.04–4.97 | 2.43 | 1.91 (±0.343) | 1.43 (±0.373) | 3.35 (±0.612) | 42.46 (±5.843) | 18.26 | 13.76 | 2.51 | 2B | 4V+36L |
| Cymbidium lowianum | 1.76–4.44 | 2.52 | 1.82 (±0.358) | 1.47 (±0.278) | 3.29 (±0.584) | 44.88 (±3.913) | 17.75 | 8.71 | 1.54 | 2B | 6V+34L |
| Cymbidium devonianum | 1.87–3.80 | 2.03 | 1.60 (±0.311) | 1.20 (±0.279) | 2.81 (±0.505) | 42.84 (±5.413) | 17.97 | 12.63 | 2.26 | 2B | 2V+38L |
| Cymbidium elegans | 1.56–4.29 | 2.75 | 1.74 (±0.436) | 1.32 (±0.331) | 3.07 (±0.672) | 43.39 (±5.352) | 21.88 | 12.33 | 2.69 | 2B | 4V+36L |
| Cymbidium aloifolium | 1.89–4.87 | 2.57 | 1.81 (±0.607) | 1.24 (±0.364) | 3.05 (±0.830) | 41.18 (±7.397) | 27.21 | 17.96 | 4.88 | 2B | 6V+34L |
| Cymbidium iridioides | 2.62–7.25 | 2.76 | 2.46 (±0.617) | 1.88 (±0.569) | 4.34 (±1.073) | 43.01 (±6.260) | 24.72 | 14.55 | 3.59 | 3B | 4V+36L |
| Cymbidium tracyanum | 2.01–8.81 | 4.38 | 3.09 (±1.462) | 2.18 (±0.782) | 5.28 (±1.462) | 41.21 (±8.030) | 27.68 | 19.48 | 5.39 | 2C | 8V+30L+2J |
Abbreviations: (SC) shortest and (LC) longest chromosome length; (p) mean long arm length; (q) mean of short chromosome length; (CL) mean of total chromosome length; (CI) mean centromeric index; (CVCL) coefficient of variation in terms of chromosome length; (CVCI) coefficient of variation in terms of centromeric index; (AI) karyotype asymmetry index.
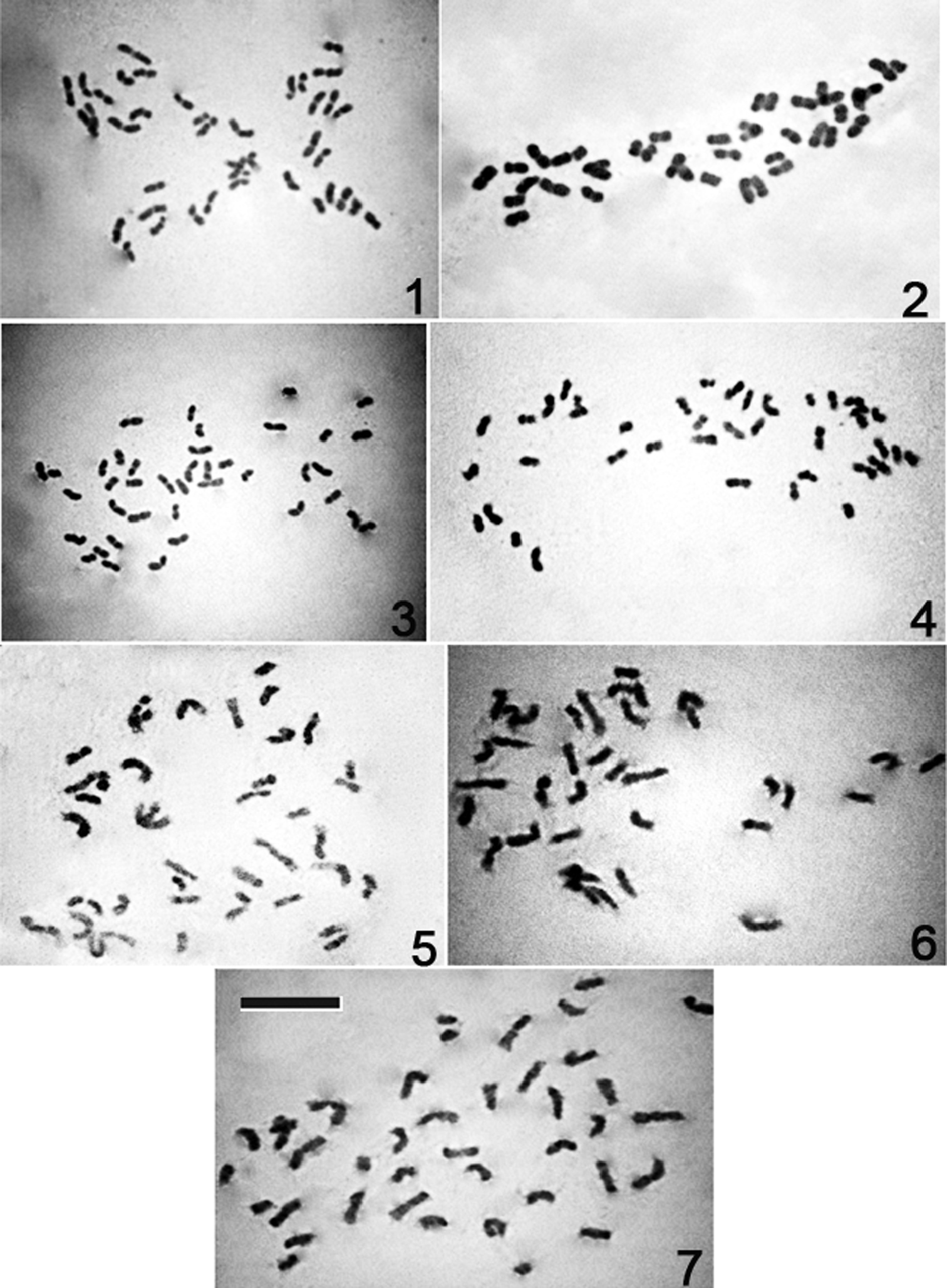
Mitotic complements of Cymbidium species. 1 Cymbidium tigrinum 2 Cymbidium lowianum 3 Cymbidium devonianum 4 Cymbidium elegans 5 Cymbidium aloifolium 6 Cymbidium iridioides 7 Cymbidium tracyanum. Bar = 10 μm.
Karyotypes of Cymbidium species. 8 Cymbidium tigrinum 9 Cymbidium lowianum 10 Cymbidium devonianum 11 Cymbidium elegans 12 Cymbidium aloifolium 13 Cymbidium iridioides 14 Cymbidium tracyanum. Heteromorphic pair marked by arrows above the short arm.
In the present study, characteristic differences have been recorded in karyotypes at inter-specific level of the genus Cymbidium. In general, nine pairs out of twenty i.e. I-II, IX-X, XIV-XVI and XVIII to XIX, showed uniformity with respect to the chromosome morphology at inter-specific level while moderate to greater degree of variation was recorded in the remaining eleven pairs of the chromosome complements pattern. Such observations indicate the high degree of gene/genome stability in the genus. In general, it is predicted that orchid seeds, which are very small and light weight, can be wind-dispersed over long distances (
The karyotypes in most of the species investigated were found to be symmetrical according to
Karyotype similarities between Cymbidium species traduce the high degree of gene stability in the genus at inter-specific level and indicate lack of chromosome structural rearrangements during speciation in Cymbidium. The present investigation may also provide useful information on chromosome markers including heteromorphic chromosomes based speciation, basic chromosome number and ploidy level vis-à-vis genome evolution; which is more or less poorly known in the family Orchidaceae and especially in the genus Cymbidium.
The present work was supported by a grant from University Grants Commission, Government of India, New Delhi, through University with Potential for Excellence (UPE) – Bioscience program. The authors are thankful to Head, Department of Biotechnology and Bioinformatics, NEHU for providing facilities. Sincere thanks are due to Prof. M.S. Bisht, Dr. C.S. Rao and all members of Plant Biotechnology Laboratories of Department of Botany and Department of Biotechnology and Bioinformatics, NEHU, Shillong, for their constant encouragement and help.