(C) 2012 Simone Lilian Gruber. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Few species of the tribe Lophiohylini have been karyotyped so far, and earlier analyses were performed mainly with standard staining. Based on the analysis of seven species with use of routine banding and molecular cytogenetic techniques, the karyotypes were compared and the cytogenetic data were evaluated in the light of the current phylogenies. A karyotype with 2n = 24 and NOR in the chromosome 10 detected by Ag-impregnation and FISH with an rDNA probe was shared by Aparasphenodon bokermanni Miranda-Ribeiro, 1920, Itapotihyla langsdorffii (Duméril and Bibron, 1841), Trachycephalus sp., Trachycephalus mesophaeus (Hensel, 1867), and Trachycephalus typhonius (Linnaeus, 1758). Phyllodytes edelmoi Peixoto, Caramaschi et Freire, 2003 and Phyllodytes luteolus (Wied-Neuwied, 1824) had reduced the diploid number from 2n = 24 to 2n = 22 with one of the small-sized pairs clearly missing, and NOR in the large chromosome 2, but the karyotypes were distinct regarding the morphology of chromosome pairs 4 and 6. Based on the cytogenetic and phylogenetic data, it was presumed that the chromosome evolution occurred from an ancestral type with 2n = 24, in which a small chromosome had been translocated to one or more unidentified chromosomes. Whichever hypothesis is more probable, other rearrangements should have occurred later, to explain the karyotype differences between the two species of Phyllodytes Wagler, 1830. The majority of the species presented a small amount of centromeric C-banded heterochromatin and these regions were GC-rich. The FISH technique using a telomeric probe identified the chromosome ends and possibly (TTAGGG)n-like sequences in the repetitive DNA out of the telomeres in Itapotihyla langsdorffii and Phyllodytes edelmoi. The data herein obtained represent an important contribution for characterizing the karyotype variability within the tribe Lophiohylini scarcely analysed so far.
Amphibian cytogenetics, Ag-NOR, C-banding, rDNA probe, telomeric probe, fluorochrome staining
The hylids of the subfamily Hylinae Rafinesque, 1815 are grouped into four large tribes: Cophomantini, Dendropsophini, Hylini, and Lophiohylini (
About 70 species are recognised in the tribe Lophiohylini (
The present paper deals with the chromosome analysis of Aparasphenodon bokermanni Pombal, 1993, Itapotihyla langsdorffii, Phyllodytes edelmoi Peixoto, Caramaschi & Freire, 2003, Phyllodytes luteolus (Wied-Neuwied, 1824), Trachycephalus mesophaeus, Trachycephalus typhonius, and Trachycephalus sp. (probably an undescribed species) with use of routine and molecular cytogenetic techniques. The aim was to analyze species never karyotyped before and to improve the cytogenetic data from some other species, in order to better characterizing the karyotype variability within the tribe Lophiohylini and to carry out a more comprehensive comparative analysis in the light of the current phylogeny.
Cytogenetic analyses were performed with specimens of Aparasphenodon Miranda-Ribeiro, 1920, Itapotihyla Faivovich, Haddad, Garcia, Frost, Campbell, et Wheeler, 2005, Phyllodytes, and Trachycephalus Tschudi, 1838 (Table 1) collected in the Brazilian states of Alagoas (AL), Bahia (BA), Espírito Santo (ES), Mato Grosso (MS), and São Paulo (SP). The voucher specimens were deposited in the amphibian collection Célio Fernando Baptista Haddad (CFBH), housed in the Departamento de Zoologia, UNESP, Rio Claro, SP, Brazil.
Species, number of individuals, sex, voucher numbers, and collection locations in Brazil.
| species | number | sex | voucher numbers CFBH | collection locations |
|---|---|---|---|---|
| Aparasphenodon bokermanni | 1 | male | 22575 | Cananéia, SP (25°01'19"S, 47°55'41"W) |
| Itapotihyla langsdorffii | 2 | males | 22369, 22370 | Ilhéus, BA (14°47'29"S, 39°02'41"W) |
| 1 | female | 30973 | Rio Claro, SP (22°25'20"S, 47°34'23"W) | |
| Phyllodytes edelmoi | 2 | females | 22583, 22584 | Maceió, AL (09°40'06"S, 35°43'59"W) |
| 1 | male | 22585 | Maceió, AL (09°40'06"S, 35°43'59"W) | |
| Phyllodytes luteolus | 2 | males | 22462, 22463 | Guaraparí, ES (20°39'01"S, 40°29'10"W) |
| Trachycephalus sp. | 1 | male | 20664 | Paranaíta, MT (09°40'56"S; 56°28'50"W) |
| Trachycephalus mesophaeus | 3 | males | 22366, 22367, 22368 | Ilhéus, BA (14°47'29"S, 39°02'41"W) |
| 2 | females | 22371, 22372 | Ilhéus, BA (14°47'29"S, 39°02'41"W) | |
| 1 | juvenile | 22484 | Ubatuba, SP (23°26'19"S, 45°05'25"W) | |
| 1 | male | 24222 | Biritiba Mirim, SP (23°34'17"S, 46°02'15"W) | |
| Trachycephalus typhonius | 1 | female | 22365 | Porto Primavera, MS (22°26'01"S, 52°58'11"W) |
| 1 | male | 10033 | Rio Claro, SP (22°25'20"S, 47°34'23"W) |
CFBH - Célio Fernando Baptista Haddad Collection, UNESP, Rio Claro, SP, Brazil.
Direct cytological suspensions of bone marrow, liver, and testes were prepared according to the procedures described in
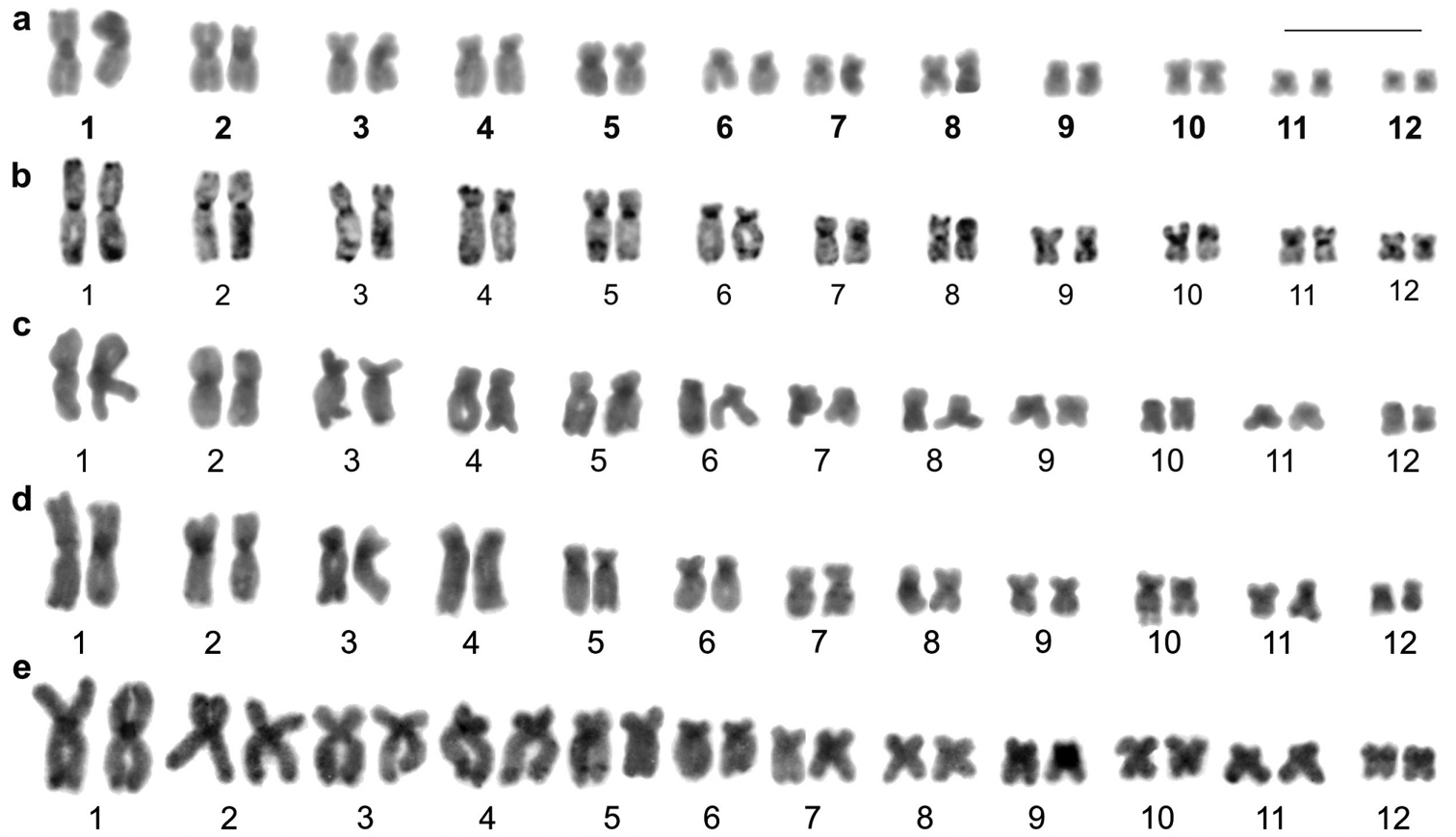
Specimens of Aparasphenodon bokermanni, Itapotihyla langsdorffii, Trachycephalus sp., Trachycephalus mesophaeus, and Trachycephalus typhonius had a diploid number of 2n = 24 (Fig. 1a–e) and a fundamental number FN = 48 and Phyllodytes edelmoi and Phyllodytes luteolus had 2n = 22, FN = 44 (Fig. 1f–g). The Table 2 presents the relative length (RL), centromeric index (CI), and the centromeric position (CP) with morphologic classification of the chromosomes of the seven species.
Giemsa-stained karyotypes. a Aparasphenodon bokermanni, male, 2n = 24 b Itapotihyla langsdorffii, male, 2n = 24 c Trachycephalus sp., male, 2n = 24 d Trachycephalus mesophaeus, male, 2n = 24 e Trachycephalus typhonius, male, 2n = 24; f. Phyllodytes edelmoi, male, 2n = 22 g Phyllodytes luteolus, male, 2n = 22. The insets show the chromosome pairs with Ag-NOR and FISH using the rDNA probe. Bar = 10 mm.
Relative length (RL), centromeric index (CI), and nomenclature for centromeric position (CP) on mitotic chromosomes according to
| Species | Chromosome number | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
| Aparasphenodon bokermanni | RL | 15.57 | 12.93 | 10.65 | 9.63 | 9.48 | 7.48 | 5.68 | 6.78 | 6.27 | 6.5 | 4.12 | 3.83 |
| CI | 0.479 | 0.459 | 0.396 | 0.263 | 0.344 | 0.286 | 0.321 | 0.464 | 0.487 | 0.284 | 0.420 | 0.465 | |
| CP | m | m | m | sm | sm | sm | sm | m | m | sm | m | m | |
| Itapotihyla langsdorffii | RL | 15.06 | 13.52 | 11.50 | 10.41 | 9.82 | 7.68 | 6.59 | 6.35 | 5.17 | 5.00 | 5.02 | 3.90 |
| CI | 0.460 | 0.421 | 0.355 | 0.241 | 0.361 | 0.225 | 0.391 | 0.483 | 0.472 | 0.472 | 0.460 | 0.467 | |
| CP | m | m | sm | st | sm | st | m | m | m | m | m | m | |
| Trachycephalus sp. | RL | 14.57 | 11.79 | 11.57 | 9.95 | 9.18 | 7.81 | 6.75 | 6.06 | 4.43 | 5.15 | 4.65 | 4.30 |
| CI | 0.430 | 0.429 | 0.383 | 0.257 | 0.319 | 0.261 | 0.344 | 0.453 | 0.456 | 0.301 | 0.443 | 0.461 | |
| CP | m | m | m | sm | sm | sm | sm | m | m | sm | m | m | |
| Trachycephalus mesophaeus | RL | 14.33 | 13.57 | 10.66 | 10.47 | 9.04 | 7.98 | 6.76 | 6.35 | 5.94 | 6.97 | 4.72 | 3.83 |
| CI | 0.457 | 0.435 | 0.366 | 0.268 | 0.370 | 0.224 | 0.338 | 0.481 | 0.424 | 0.351 | 0.353 | 0.414 | |
| CP | m | m | sm | sm | sm | st | sm | m | m | sm | sm | m | |
| Trachycephalus typhonius | RL | 15.63 | 12.80 | 11.05 | 10.51 | 10.06 | 8.16 | 7.07 | 6.02 | 5.02 | 5.23 | 4.59 | 4.10 |
| CI | 0.462 | 0.397 | 0.364 | 0.236 | 0314 | 0.200 | 0.317 | 0.424 | 0.444 | 0.304 | 0.461 | 0.485 | |
| CP | m | m | sm | st | sm | st | sm | m | m | sm | m | m | |
| Phyllodytes edelmoi | RL | 18.38 | 13.74 | 12.88 | 9.90 | 9.74 | 7.73 | 6.86 | 6.77 | 4.88 | 4.06 | 3.74 | -- |
| CI | 0.453 | 0.403 | 0.335 | 0.430 | 0.341 | 0.414 | 0.367 | 0.404 | 0.440 | 0.444 | 0.472 | -- | |
| CP | m | m | sm | m | sm | m | sm | m | m | m | m | -- | |
| Phyllodytes luteolus | RL | 16.62 | 12.56 | 11.11 | 10.65 | 9.57 | 8.72 | 8.38 | 7.05 | 5.38 | 4.80 | 4.56 | -- |
| CI | 0.450 | 0.422 | 0.370 | 0.249 | 0.352 | 0.237 | 0.336 | 0.354 | 0.472 | 0.430 | 0.443 | -- | |
| CP | m | m | sm | st | sm | st | sm | sm | m | m | m | -- | |
m = metacentric; sm = submetacentric; st = subtelocentric.
The technique of Ag-NOR was carried out in all species. In the 2n = 24 karyotypes the Ag-NORs were located on chromosome 10, at the terminal long arm in the case of Aparasphenodon bokermanni, Trachycephalus sp., Trachycephalus mesophaeus, and Trachycephalus typhonius (Fig. 1a, c–e), or at the interstitial short arm in Itapotihyla langsdorffii (Fig. 1b). In Phyllodytes edelmoi and Phyllodytes luteolus Ag-NOR was located at the terminal long arm of chromosome 2 (Fig. 1f–g). The Ag-impregnation occurred in the sites of the secondary constriction, although this marker was not always visualised in the standard stained chromosomes. In Aparasphenodon bokermanni and Trachycephalus sp. and in some individuals of the remaining species, there was variation in the pattern of Ag-NOR labelling. Within the same individual, metaphases exhibited Ag-NORs with conspicuous or slight difference in size or carried two Ag-NORs with equivalent sizes; occasionally a single Ag-NOR per metaphase was also observed in the same cytological preparation. FISH with an rDNA probe was performed in six species, with exception of Phyllodytes edelmoi. Two fluorescent signals were observed in all analysed metaphases (Fig. 1a–e, g). In the species Trachycephalus sp. and Trachycephalus mesophaeus the hybridisation signals always presented the same size and in Aparasphenodon bokermanni, Itapotihyla langsdorffii, Trachycephalus typhonius, and Phyllodytes luteolus the labelling was heteromorphic in all metaphases.
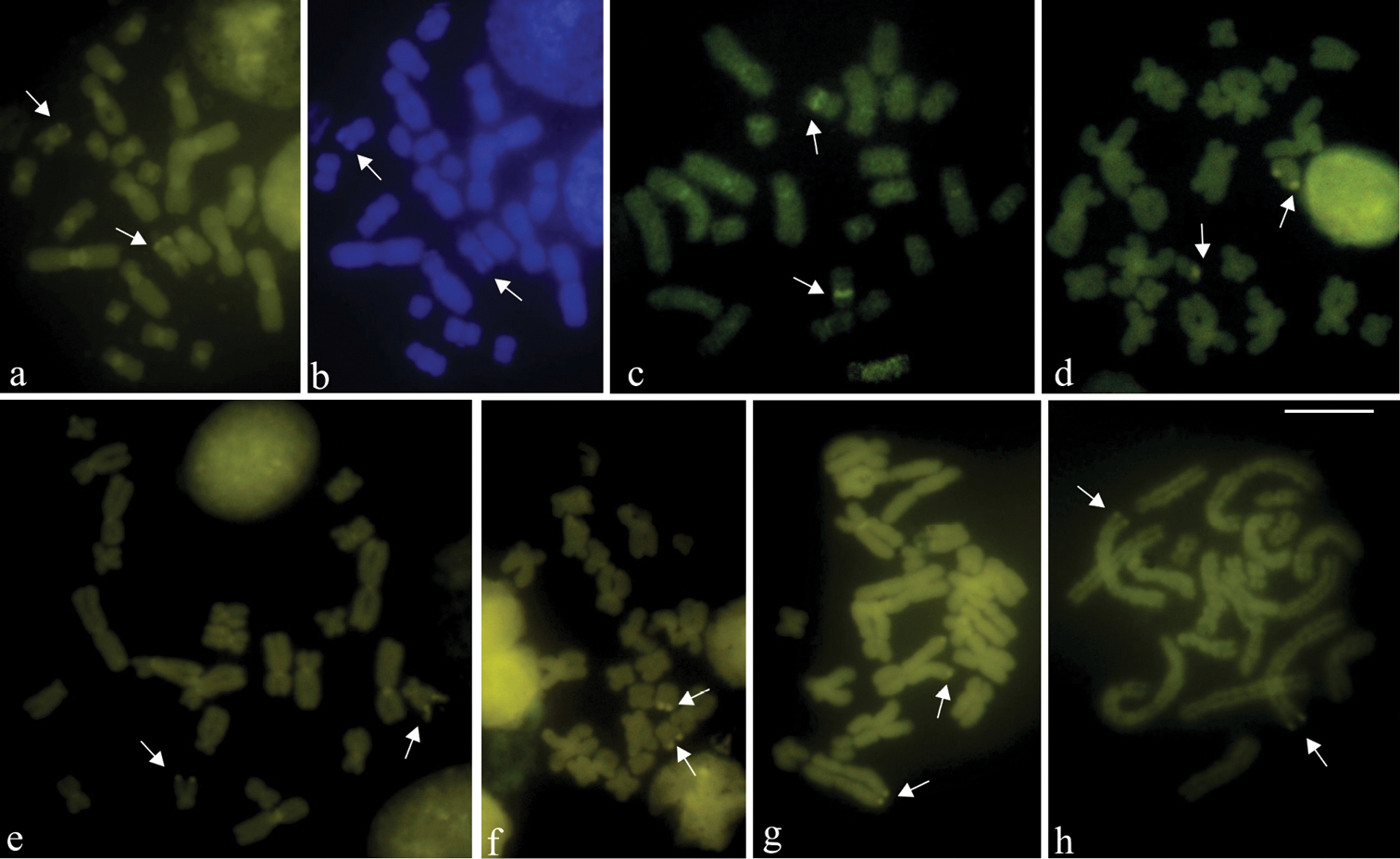
The C-banding in Aparasphenodon bokermanni, Itapotihyla langsdorffii, Trachycephalus sp., Trachycephalus mesophaeus, and Trachycephalus typhonius showed heterochromatin distribution in the pericentromeric regions of all chromosomes (Fig. 2). In Itapotihyla langsdorffii additional C-bands were noticed at terminal (chromosomes 1 and 4) and interstitial (chromosome 5) regions. This technique was carried out in mitotic and meiotic cytological preparations of Phyllodytes edelmoi and Phyllodytes luteolus, but no C-banded region was demonstrated in the chromosomes of these species. The NOR site in all species was brilliant with CMA3, as well as the chromosome pericentromeric region (Fig. 3a, c–h). The pericentromeric fluorescence was in general faint and not visualised in all chromosomes. In Aparasphenodon bokermanni the centromeric signals were particularly prominent in size and brightness (Fig. 3a). No brilliant labelling was observed after DAPI staining in any species, except in Aparasphenodon bokermanni which showed slight fluorescence at the terminal short arm of chromosome 10 (Fig. 3b). The chromosome pericentromeric region of this species was DAPI-negative.
C-banded karyotypes. a Aparasphenodon bokermanni b Itapotihyla langsdorffii c Trachycephalus sp. d Trachycephalus mesophaeus e Trachycephalus typhonius. Bar = 10 mm.
Fluorochrome-stained metaphases. a, c-h CMA3b DAPI a–b Aparasphenodon bokermanni c Itapotihyla langsdorffii d Trachycephalus sp. e Trachycephalus mesophaeus f Trachycephalus typhonius g Phyllodytes edelmoi h Phyllodytes luteolus. Bright DAPI fluorescence at the terminal short arms of chromosomes 10 (arrows) and the negative centromeric region are shown in a. CMA3 fluorescent labelling of the NOR site (arrows) and in the centromeric region of chromosomes in a, c–h. Bar = 10 mm.
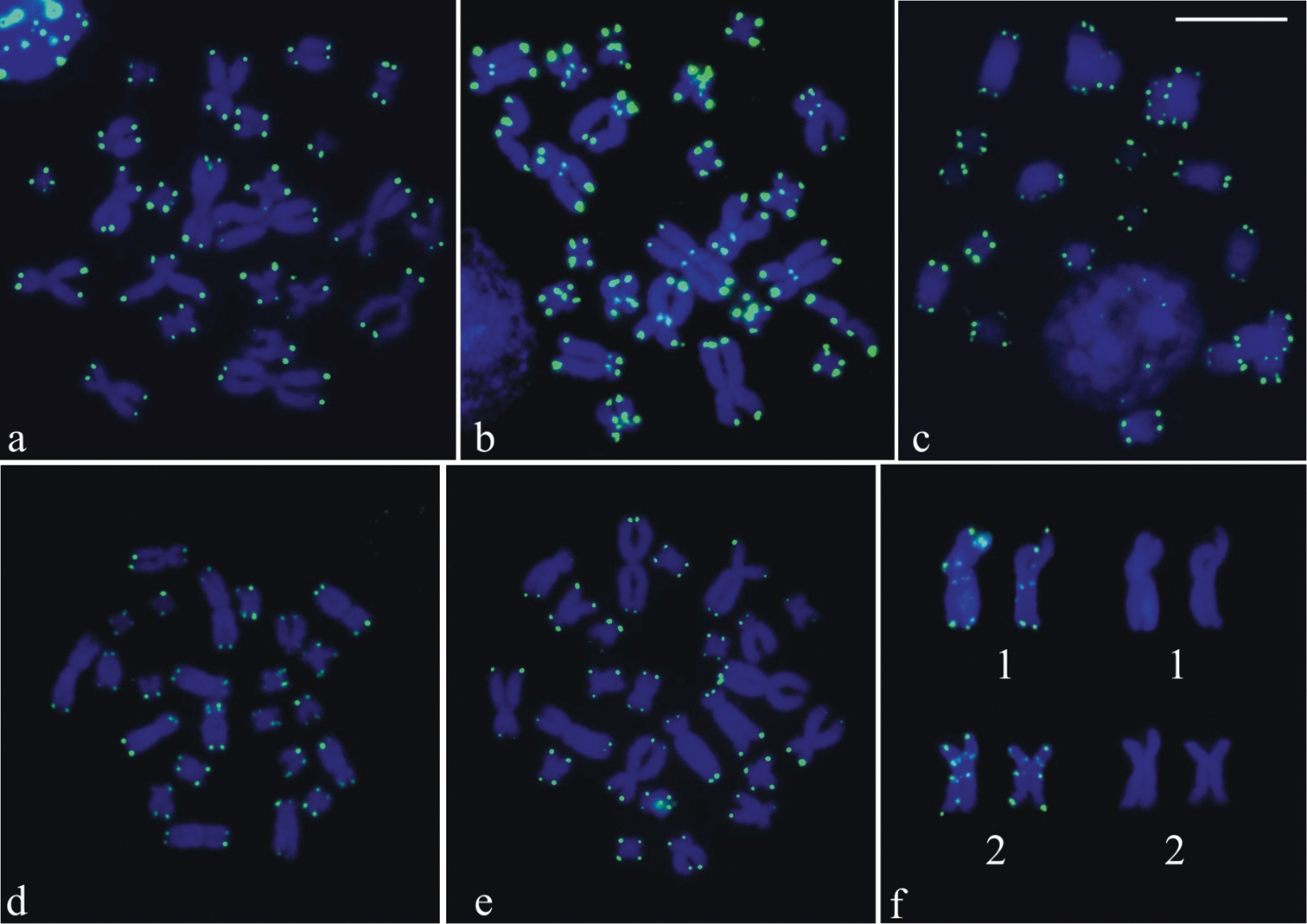
The telomeric probe hybridized on the chromosome ends in six of the species, excepting in Phyllodytes luteolus without cytological material available for the FISH technique. Figure 4a–e showed metaphases of Aparasphenodon bokermanni, Itapotihyla langsdorffii, Trachycephalus sp., Trachycephalus mesophaeus, and Trachycephalus typhonius with probe labelling at the chromosome ends and, in the case of Itapotihyla langsdorffii (Fig. 4b), also in the pericentromeric region. In Phyllodytes edelmoi no good metaphases were obtained, but the chromosomes showed telomeric labelling. In one metaphase of this species, however, the large-sized chromosome pair 1 and 2 had probe hybridization at the proximal short and long arms (Fig. 4f).
FISH using a telomeric probe. a Aparasphenodon bokermanni b Itapotihyla langsdorffii c Trachycephalus sp. d, Trachycephalus mesophaeus e Trachycephalus typhonius f Phyllodytes edelmoi. In b hybridisation labelling is visible in the centromeric region of the chromosomes and in f, at the proximal short and long arms of chromosomes 1 and 2 observed with telomeric probe hybridisation (left) and with DAPI staining (right). Bar = 10 mm.
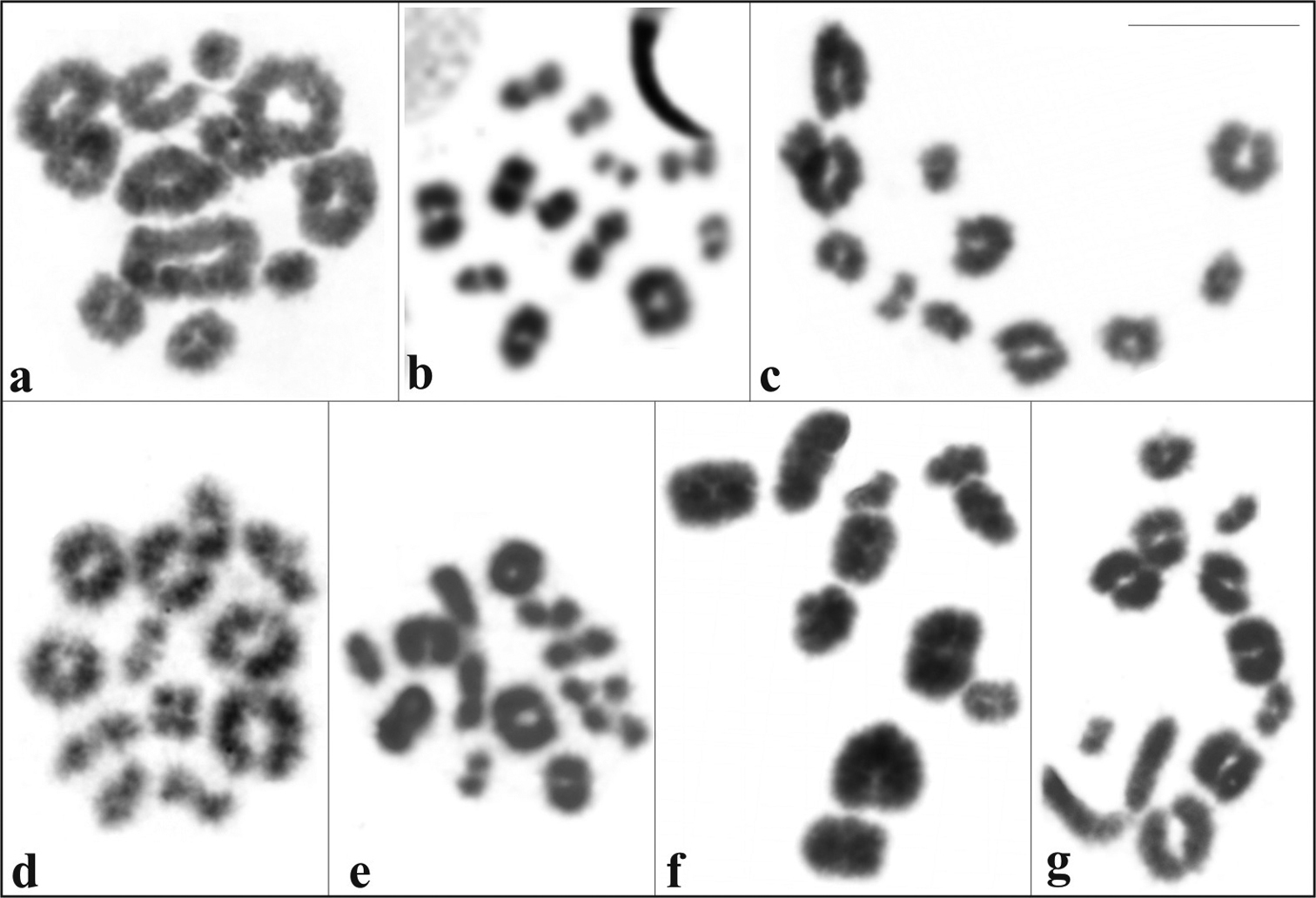
No sex-chromosome pairs were detected in male or female specimens of Itapotihyla langsdorffii, Trachycephalus mesophaeus, Trachycephalus typhonius, and Phyllodytes edelmoi. In the remaining three species only males were karyotyped with no evidence of sex related heteromorphism. Meiotic analysis confirmed the diploid number in all species (Fig. 5a–g). Aparasphenodon bokermanni, Itapotihyla langsdorffii, Trachycephalus sp., Trachycephalus mesophaeus, and Trachycephalus typhonius showed 12 bivalents. Phyllodytes edelmoi and Phyllodytes luteolus showed 11 bivalents.
Giemsa-stained diakinesis and metaphases I cells. a Aparasphenodon bokermanni, 2n = 24 b Itapotihyla langsdorffii, 2n = 24 c Trachycephalus sp., 2n = 24 d Trachycephalus mesophaeus, 2n = 24 e Trachycephalus typhonius, 2n = 24 f Phyllodytes edelmoi, 2n = 22 g Phyllodytes luteolus, 2n = 22. Bar = 10 mm.
The main cytogenetic data obtained in the present study are summarized in the Table 3.
Data on chromosome number, chromosome formula, NOR and telomeric sequence localization, C-band distribution and molecular content of repetitive DNA sequences of studied species.
| species | 2n | fomula | NOR | Tel | C bands | DAPI | CMA3 |
|---|---|---|---|---|---|---|---|
| Aparasphenodon bokermanni | 24 | 7m+5sm | 11qt | T | C+NOR | 10pt | C*+NOR |
| Itapotihyla langsdorffii | 24 | 8m+2sm+2st | 11pi | T+C | C+NOR | -- | C+NOR |
| Trachycephalus sp. | 24 | 7m+5sm | 11qt | T | C+NOR | -- | C+NOR |
| Trachycephalus mesophaeus | 24 | 5m+6sm+1st | 11qt | T | C+NOR | -- | C+NOR |
| Trachycephalus typhonius | 24 | 6m+4sm+2st | 11qt | T | C+NOR | -- | C+NOR |
| Phyllodytes edelmoi | 22 | 8m+3sm | 2qt | T+C | C+NOR | -- | C+NOR |
| Phyllodytes luteolus | 22 | 5m+4sm+2st | 2qt | --- | C+NOR | -- | C+NOR |
m = metacentric; sm = submetacentric; st = subtelocentric; p = short chromosome arm; q = long chromosome arm; i = interstitial region; t = terminal region; T = telomere; C = centromeric/ pericentromeric region; * intense mark.
The species of the tribe Lophiohylini Aparasphenodon bokermanni, Itapotihyla langsdorffii, Trachycephalus sp., Trachycephalus mesophaeus, and Trachycephalus typhonius with 2n = 24 shared indistinguishable karyotypes even though there was discrepancy in morphological classification shown in Table 2 for some chromosomes, as the chromosome 3 of the species, due to slight differences in the chromosome arm proportion. No evidence of population karyotype difference was observed for Itapotihyla langsdorffii, Trachycephalus mesophaeus, and Trachycephalus typhonius sampled in distinct locations. Considering previous data for these three species (
The chromosome constitution with 2n = 24 herein described is the same as found for the remaining eight species of Lophiohylini analysed so far, corresponding to Aparasphenodon brunoi, Argenteohyla siemersi, Corythomantis greeningi, Osteocephalus taurinus, Osteopilus dominicensis, Osteopilus marianae, Osteopilus septentrionalis, and an unidentified species of Trachycephalus (see
Phyllodytes edelmoi and Phyllodytes luteolus, the first two species of the genus that were analysed so far, had reduced the diploid numbers from 2n = 24 to 2n = 22 and the NOR site was in the large-sized chromosome 2. Nevertheless, the karyotypes of these two species were distinct regarding the morphology of pairs 4 and 6, that is, in Phyllodytes edelmoi these pairs were metacentric and in Phyllodytes luteolus they were subtelocentric, as it has been usually observed in Hylinae species with 2n = 24. The discrepancy in the morphology of the chromosome pairs 4 and 6 was supported by the chromosome measurements and the mechanism responsible for these differences might be, for example, a pericentric inversion or another type of chromosome rearrangement, but this could not be determined at least with the cytogenetic techniques used here.
Within the sub-family Hylinae, variation as resulted of fusion events from an ancestral karyotype with 24 chromosomes was described for Hypsiboas albopunctatus (Spix, 1824) (2n = 22) and for species of the genus Aplastodiscus (2n = 18, 20, 22) (
In males and females of Itapotihyla langsdorffii, Phyllodytes edelmoi, T mesophaeus, and Trachycephalus typhonius and in males of Aparasphenodon bokermanni, Trachycephalus sp., and Phyllodytes luteolus heteromorphic sex chromosomes were not observed. Nevertheless in females of these three latter species sex chromosomes could not be discarded. Anurans, in general, do not present cytological sex chromosome differentiation and both male or female heterogamety has been described in some species (
A single NOR pair located in a small-sized chromosome (
The NOR marker chromosome in our species of Lophiohylini with 2n = 24 was considered as the 10 and the rDNA sequences were at the interstitial short arm or at the terminal long arm, but no major differences were observed in the morphology of the chromosomes 10 among distinct species. Therefore, the mechanism that changed the NOR sites apparently was not a gross rearrangement, but minute structural rearrangements or transposition by means of mobile elements could not be discarded. If the movement of the NOR from chromosome 10 to chromosome 2 in Phyllodytes species was not a direct consequence of the rearrangement which reduced the diploid number in the genus, one of the two mentioned mechanisms would also be a reasonable explanation for the discrepant NOR site, in Phyllodytes edelmoi and in Phyllodytes luteolus.
The technique of Ag-impregnation showed large variation in the Ag-NOR pattern within the same individual. Nevertheless, the FISH with an rDNA probe revealed that the NOR labelling in each individual had either equivalent or distinct size in all the analysed cells. Such data allowed us to conclude that most probably the Ag-NOR variation was a result of differential activity of ribosomal gene in Trachycephalus sp. and Trachycephalus mesophaeus because the hybridization labelling had the same size in both homologues; on the other hand, different amounts of repetitive rDNA units would be responsible for the observed Ag-NOR variation in Aparasphenodon bokermanni, Itapotihyla langsdorffii, and Trachycephalus typhonius because hybridization labelling had distinct sizes in both homologues. The single Ag-NOR seen occasionally in some metaphases could be attributed to the lacking or insufficient amount of the non-histone proteins available for the Ag-impregnation.
The chromosomes of the species herein analysed produced C-banding results only after over treatment of the distinct steps of the technique. However, it was undoubtedly demonstrated that heterochromatin was distributed mainly in the centromeric regions. A similar centromeric C-banding pattern had been described in Aparasphenodon brunoi, Corythomantis greeningi, and Itapotihyla langsdorffii (
Surprisingly, in spite of the low amount of C-banded heterochromatin, Aparasphenodon bokermanni showed conspicuous bright fluorescence at the centromeres, similar to that observed in Aparasphenodon brunoi (
Although the FISH with the telomeric probe is primarily designed for identification of chromosome-ends, this procedure may provide information about the molecular nature of some repetitive sequences. As far as it has been shown, distinct organisms, including frogs (
The interstitial hybridization signals of telomeric probe could correspond to vestiges of true telomeres, as reported in rodents (
Based on the data of 22 species, a phylogenetic tree of the Lophiohylini was provided by
The present study showed that in spite of the high similarity of the chromosome constitution and of the NOR pattern among the species of Lophiohylini with 2n = 24, the karyotypes could be recognized by the nature of the repetitive sequences, as differentiated through C-banding, base-specific fluorochrome staining, and, in a certain extension, by FISH with telomeric probe. Cytogenetic information on the tribe is still minimal, but the analyses of the available data in light of the phylogeny allowed for visualization of the occurrence of karyotypic variations restricted to the clades of the genera Phyllodytes and Osteopilus. It would be interesting to enlighten the chromosome evolution with other accurate technical approaches and to extend the karyotyping to other species of Lophiohylini, especially new representatives of Phyllodytes and Phytotriades auratus.
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). The authors thank to Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) for providing the collection permits to SLG and CFBH. The authors are grateful to Akio Miyoshi, Carlos Jared, Edson Zefa, Hideki Narimatsu, João Luiz Gasparini, Juliana Zina, Katyuscia de Araujo Vieira, and Olívia Araújo, for help during field work.