(C) 2012 Uedson Pereira Jacobina. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
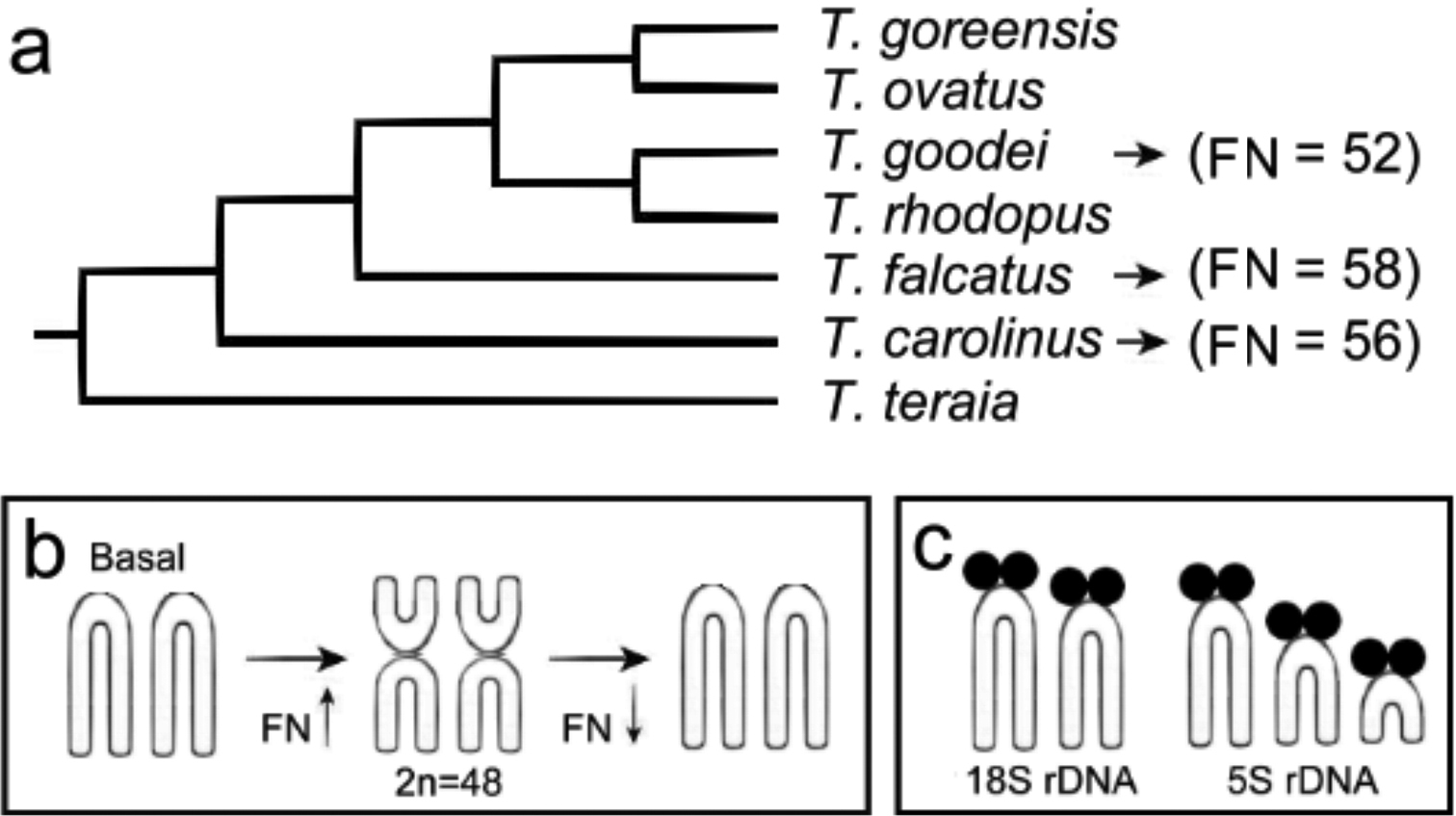
Chromosomal traits have provided valuable information for phylogeny and taxonomy of several fish groups. Three Atlantic Carangidae species of the genus Trachinotus Lacépède, 1801 (Trachinotus goodei Jordan et Evermann, 1896, Trachinotus carolinus (Linnaeus, 1766)and Trachinotus falcatus (Linnaeus, 1758)) were investigated, having 2n=48 chromosomes but different chromosomal arms (FN number), i.e., 52, 56 and 58, respectively, in view of the different number of two-armed chromosomes found in their karyotypes. Thus, Trachinotus goodei, Trachinotus carolinus and Trachinotus falcatus present a progressive distancefrom the probable basal karyotype proposed for Perciformes (2n=48 acrocentrics, FN=48). At first sight, these findings do not agree with the phylogenetic hypothesis based on mitochondrial sequences, where Trachinotus goodei appear as the most derived species, followed by Trachinotus falcatus and Trachinotus carolinus, respectively. However, the chromosomal mapping of ribosomal DNAs was informative for clarifying this apparent conflict. Indeed, the multiple 5S and 18S rDNA sites found in Trachinotus goodei corroborate the most derived condition for this species. In this sense, the occurrence of the unexpected number of two-armed chromosomes and FN value for this species, as well as for Trachinotus carolinus, must be due to additional rounds of acrocentric formation in these species, modifying the macrostructure of their karyotypes.
Carangidae, 18S rDNA, 5S rDNA, cytotaxonomic markers, evolutionary pathways
The genus Trachinotus Lacépède, 1801, also known as pompanos, encompasses 20 species distributed in tropical and subtropical oceans (
In addition to their biological significance in commercial and sport fishing, representatives of the genus Trachinotus are considered potentially suitable for pisciculture purposes (
Most species of the marine Perciformes exhibit a basal karyotype composed of 2n=48 acrocentric chromosomes, extensively conserved in several families (
Samples of the species Trachinotus carolinus (N=5; 3 males. one female, one immature), Trachinotus falcatus (N=10; 4 males, 3 females, 3 immatures) and Trachinotus goodei (N=10; 6 males, 4 females) were obtained on the coast of Rio Grande do Norte state (05°05'26"S, 36°16'31"W), in Northeast Brazil. Prior to chromosomal preparations, specimens were submitted to in vivo mitotic stimulation for 24 hours, through intramuscular and intraperitoneal injection of complex antigens (
Chromosomes were classified as metacentric (m), submetacentric (sm), subtelocentric (st) and acrocentric (a), based on the system proposed by
The heterochromatic and nucleolar organizer regions (Ag-NORs) were identified using techniques developed by
Two probes were used: an 18S rDNA probe obtained from the nuclear DNA of Prochilodus argenteus Spix et Agassiz, 1829 (
The overall hybridization procedure followed the protocol described by
All species analyzed exhibited 2n=48 chromosomes, however with a notable difference in the number of two-armed (bibrachial) elements.
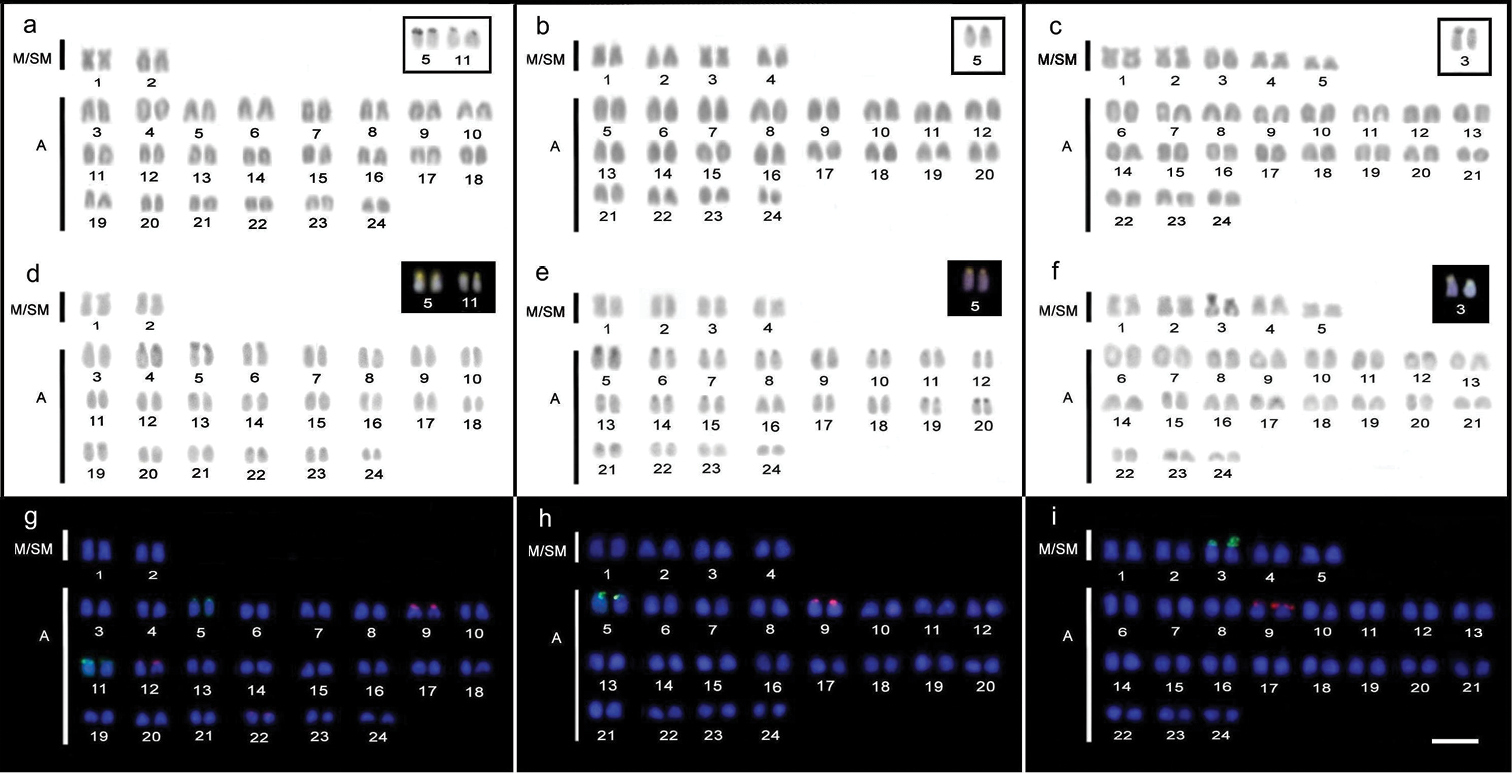
The karyotype of Trachinotus goodei (Figure 1a, d, g) is composed of 4 m/sm and 44a (FN=52). The heterochromatic regions in this species are very reduced and restricted to small blocks in the chromosomal pericentromeric regions. The Ag-NORs/18S rDNA sites were identified near the centromeric region of two acrocentric pairs, tentatively No. 5 and 11 of the karyotype. These sites proved to be rich in GC base composition (CMA+/DAPI-) (Figure 1d). Hybridization signals with 5S rDNA probes were also identified on the terminal regions of the short arms of three acrocentric pairs, tentatively numbered as 9, 12 and 22 (Figure 1g).
The Trachinotus carolinus karyotype (Figures 1 b, e, h) consists of 8m/sm and 40a (FN=56). The content of heterochromatin is also poorly distributed in the pericentromeric regions of some chromosome pairs. Ag-NORs/18S rDNA sites were located on the short arm of only one acrocentric pair, identified as number 5. These sites are clearly heterochromatic, with a CMA+/DAPI- pattern. The 5S rDNA sites were mapped only on the short arm of the acrocentric chromosome 9.
The karyotype of Trachinotus falcatus (Figure 1c, f, i) has the largest number of bibrachial elements if compared to the other species, i.e., 10 m/sm and 38a (FN=58). As in the two previous species, small heterochromatic blocks are present in pericentromeric regions of the chromosomes. Ag-NORs/18S rDNA sites were situated in the terminal region of the short arm of the submetacentric chromosome pair 3, which also appears heterochromatic after C-banding, with a CMA+/DAPI- pattern. The 5S rDNA sites were mapped exclusively on the short arms of the acrocentric pair 9.
Karyotypes of Trachinotus goodei (a, d, g), Trachinotus carolinus (b, e, h) and Trachinotus falcatus (c, f, i). Conventional staining (a, b, c) highlighting the chromosomal pairs carrying Ag-NOR sites; C-banding (d, e, f); nucleolar organizer pairs are highlighted by staining with CMA3+/DAPI-. Dual-colorFISH (f, g, h) showing the chromosomal mapping of the 18S rDNA (green) and 5S rDNA (red) sites. Bar = 5 µm.
As in many species of Perciformes, the species analyzed displayed 2n=48 and large numbers of acrocentric chromosomes, although there were notable differences in karyotype macrostructure. This is particularly evident for the number of chromosome arms (FN) that varies between species. Thus, Trachinotus goodei exhibits FN=52, Trachinotus carolinus FN=56 and Trachinotus falcatus FN=58. Karyotypes similar to those presented here for Trachinotus goodei and Trachinotus falcatus were previously identified in other populations of this species on the Southeast and Northeast coasts of Brazil (
Evolutionary karyotype modifications resulting from pericentric inversions are common in Perciformes. In fact, two-armed chromosomes have been found in approximately 30% of Carangidae species karyotyped to date (
Basing on morphological and molecular evidences, the genus Trachinotus is included in the tribe Trachinotini, which is considered one of the least diverse groups among carangids (
Whereas the fully acrocentric karyotype with 2n=48 (FN=48) is considered basal for Perciformes, variations of this karyotypic formula can be interpreted as derived conditions. Thus, an increase in the number of two-armed chromosomes, as sequentially found in Trachinotus carolinus (eight two-armed chromosomes) and in Trachinotus falcatus (ten two-armed chromosomes), would be expected to represent derived cytogenetic characteristics. As such, Trachinotus goodei, showing only four two-armed chromosomes and, consequently, the largest number of acrocentric chromosomes, would be representing the species with the karyotype closer to the basal one.
Many closely related species of Perciformes show poorly varied or cryptic cytogenetic characteristics, hampering their application in phylogenetic inferences (
Simple ribosomal sites are considered an ancestral condition, most frequently found in carangids (
Variations in the number and location of NORs in some cases, are likely to be favored by a high and heterogeneous heterochromatic content, whereas the inverse seems to reduce the evolutionary dynamism of these regions (
The existing set of cytogenetic data for Carangidae suggests karyotype evolution strongly mediated by pericentric inversion events. Based on the basal karyotype for Perciformes (2n=48 acrocentrics, FN=48), the increase of FN indicates a derived condition. Thus, if Trachinotus goodei is the most derived species in respect to Trachinotus falcatus and Trachinotus carolinus, as indicated by mitochondrial sequences (
Our understanding of the karyotype evolution of Carangidae (including rDNA) was improved by the present findings. Our data demonstrate that, in addition to structural changes by pericentric inversions, rDNA sequences also acted as an important evolutionary indicator in Trachinotus karyotype. In this sense, the combined mapping of 18S and 5S rDNA sequences proved to be useful to clarify the relationships in this fish group.
Phylogenetic tree from molecular data of some species of Trachinotini tribe (a), adapted from
We are grateful to the Coordination for the Improvement of Higher Education Teaching Personnel (CAPES) and the National Council for Scientific and Technological Development (CNPq) (Project No. 556793/2009-9) for their financial support. We also thank IBAMA (Process No. 19135/1) and José Garcia Júnior for the taxonomic identification of the species.