(C) 2013 G. Venu. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Uraeotyphlus oxyurus (Dumeril et Bibron, 1841), Uraeotyphlus interruptus Pillai et Ravichandran, 1999, Uraeotyphlus narayani Seshachar, 1939 and Uraeotyphlus menoni Annandale, 1913 were cytogenetically analysed following conventional and differential staining techniques. These species show similar karyotypes with 2n=36 (FN=58). There were no traces of species-specific features in regard to C-banding and NOR staining. The comparative study of karyotypes shows chromosomal homologies among the four species. Chromosomal data seem to support the concept that two species groups exist in the genus Uraeotyphlus.
Uraeotyphlus oxyurus species group , karyotypes, chromosomal homology
The genus Uraeotyphlus Peters, 1879is endemic to the Western Ghats region of peninsular India and constitutes one of the three genera within the family Ichthyophiidae Taylor, 1968 along with Caudacaecilia Taylor, 1968 and Ichthyophis Fitzinger, 1826(
On the basis of morphological features such as cylindrical body, annulation and of the presence or absence of phallodeum, and of limited molecular evidence,
Earlier,
In this study, we present the karyotypes of Uraeotyphlus oxyuryus and Uraeotyphlus interruptus and the results of reanalysis of chromosomes of Uraeotyphlus narayani and Uraeotyphlus menoni with a view to providing new insights into the intragenus relationships within the genus Uraeotyphlus.
Specimens of both sexes, collected from different regions the Western Ghats (Table 1) a few days before the experiment, were kept in glass aquaria under suitable conditions. After in vivo colchicine treatment, chromosome preparations were obtained from the liver, the gut epithelium and the testis. Cell suspensions, hypotonic treatment and fixation of cells were performed as described earlier (
Conventional C-banding was performed according to
Location of nucleolus organizer regions (NORs) was performed by applying the one-step silver nitrate method of
Details of collection of Uraeotyphlus interruptus, Uraeotyphlus narayani, Uraeotyphlus menoni and Uraeotyphlus oxyurus
| Species | Locality | Habitat | Voucher number | No. of animals used | Geographical coordinates |
|---|---|---|---|---|---|
| Uraeotyphlus interruptus | Gudalur, Nilgiris (Dt), Tamil Nadu, India | Mixed plantations of tea, banana, pepper, orange, coffee | BUB114, 103BUB105, 111 | 2 males2 females | 11°30'0"N, 76°30'00"E |
| Uraeotyphlus narayani | Changanssery, Kottayam (Dt), Kerala, India | Backyard garden with banana plantation | BUB101, 115BUB109, 116 | 2 males2 females | 9°28'00"N, 76°33'00"E |
| Uraeotyphlus menoni | Mattathur, Thrissur (Dt), Kerala, India | Backyard garden with banana plantation | BUB107, 113BUB106, 102 | 2 males2 females | 10°22'45"N, 76°19'15"E |
| Uraeotyphlus oxyurus | Agali, Palakkad (Dt), Kerala, India | Cultivated agricultural land with banana and coconut plantation | BUB104, 112BUB108, 110 | 2 males2 females | 11°5'0"N, 76°35'0"E |
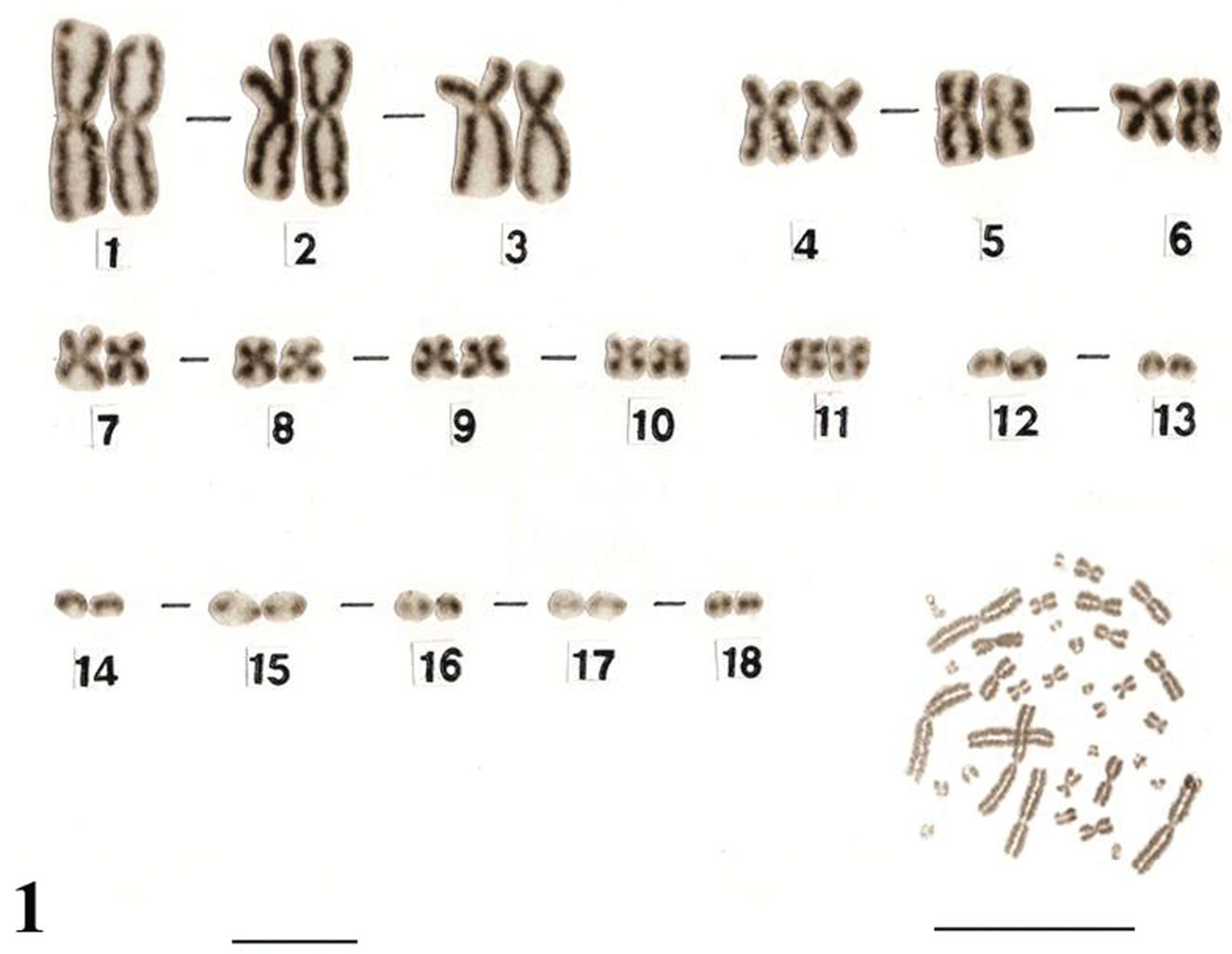
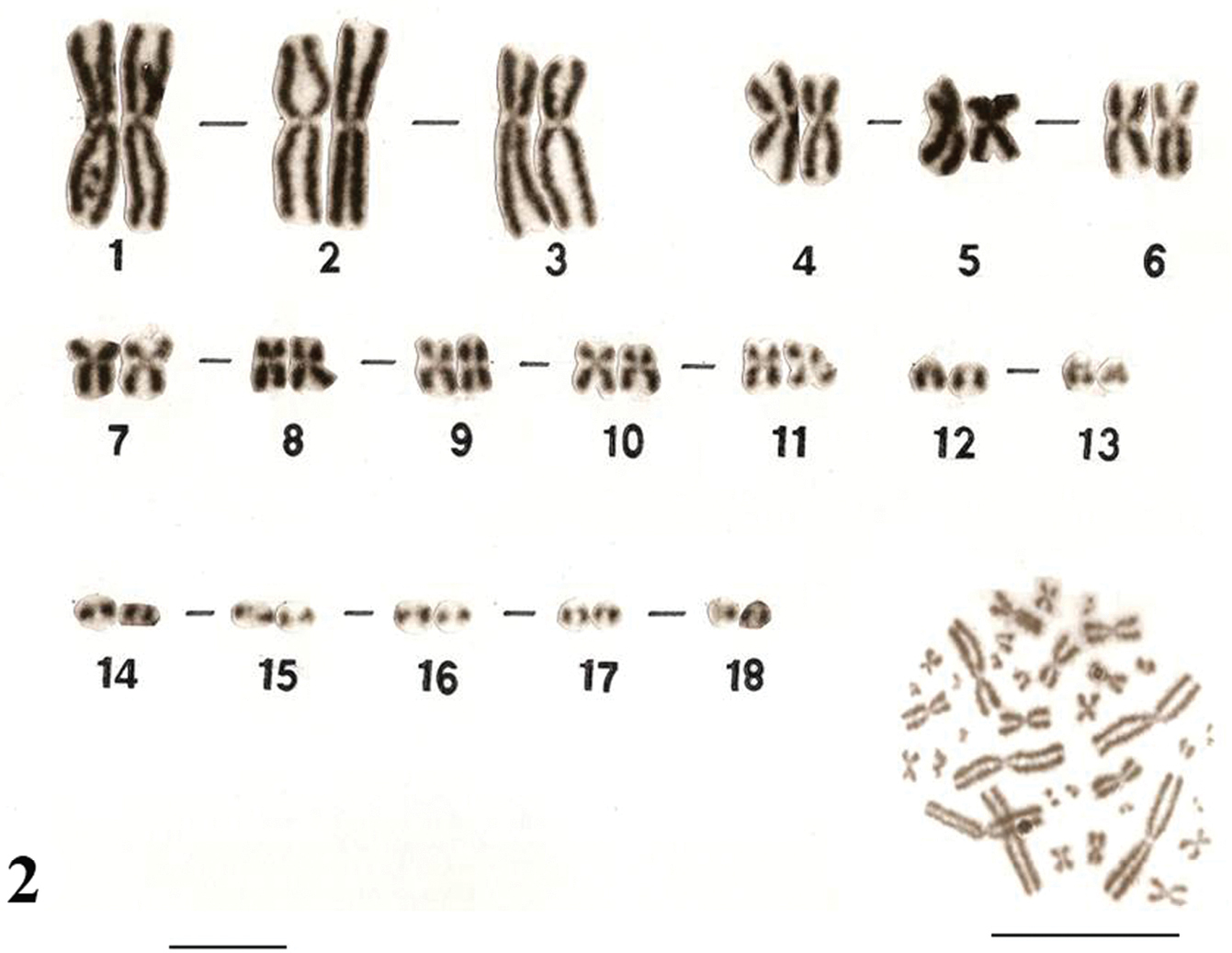
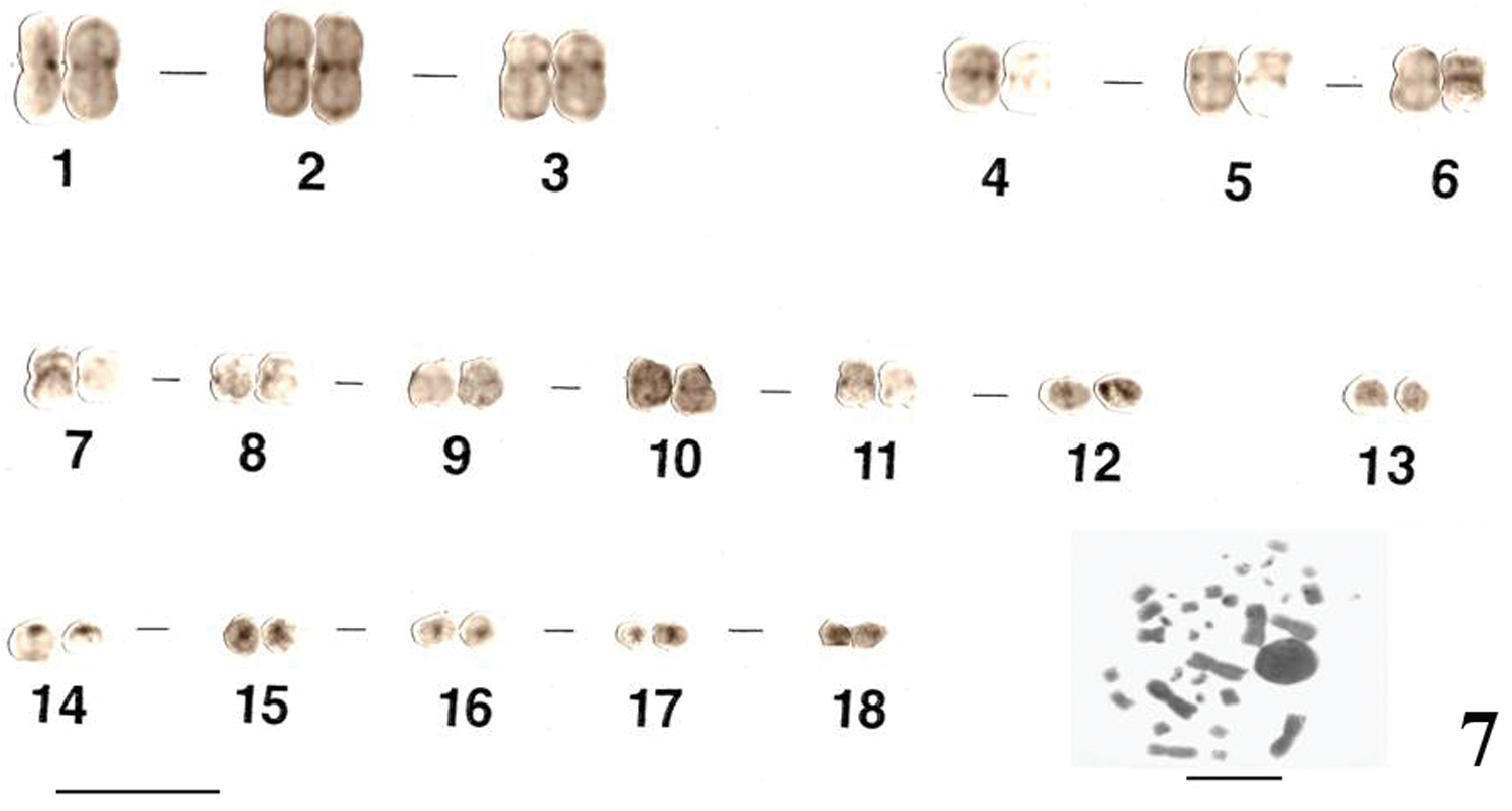
The karyotypes of Uraeotyphlus oxyurus (Fig. 1) and Uraeotyphlus interruptus (Fig. 2) revealed a diploid chromosomal complement consisting of 2n=36, FN=58.
The somatic metaphase chromosomes in the karyotype could be divided into four groups, A, B, C and D, based on the decreasing order of total length and position of centromere of each chromosome. The first group (A) includes two pairs (1–2) of large metacentrics and one pair (3) of submetacentrics, while the B group consisted of three pairs (4–6) of medium sized metacentrics in which the pair four was slightly longer than other two pairs. The third group (C) included 5 pairs (7–11) of smaller submetacentrics, all in decreasing order of their total length. The fourth group (D) included mostly acrocentric (12–18) pairs.
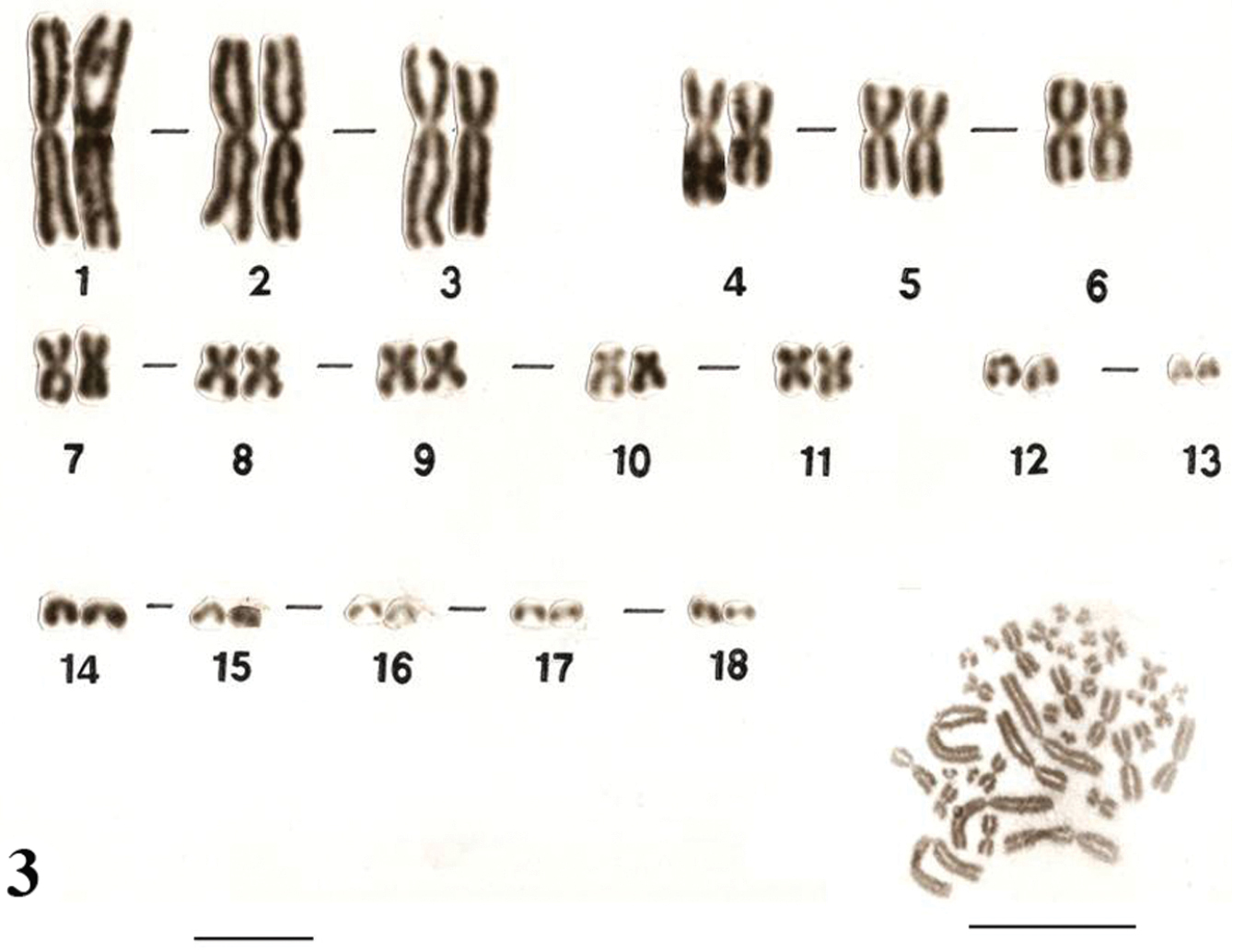
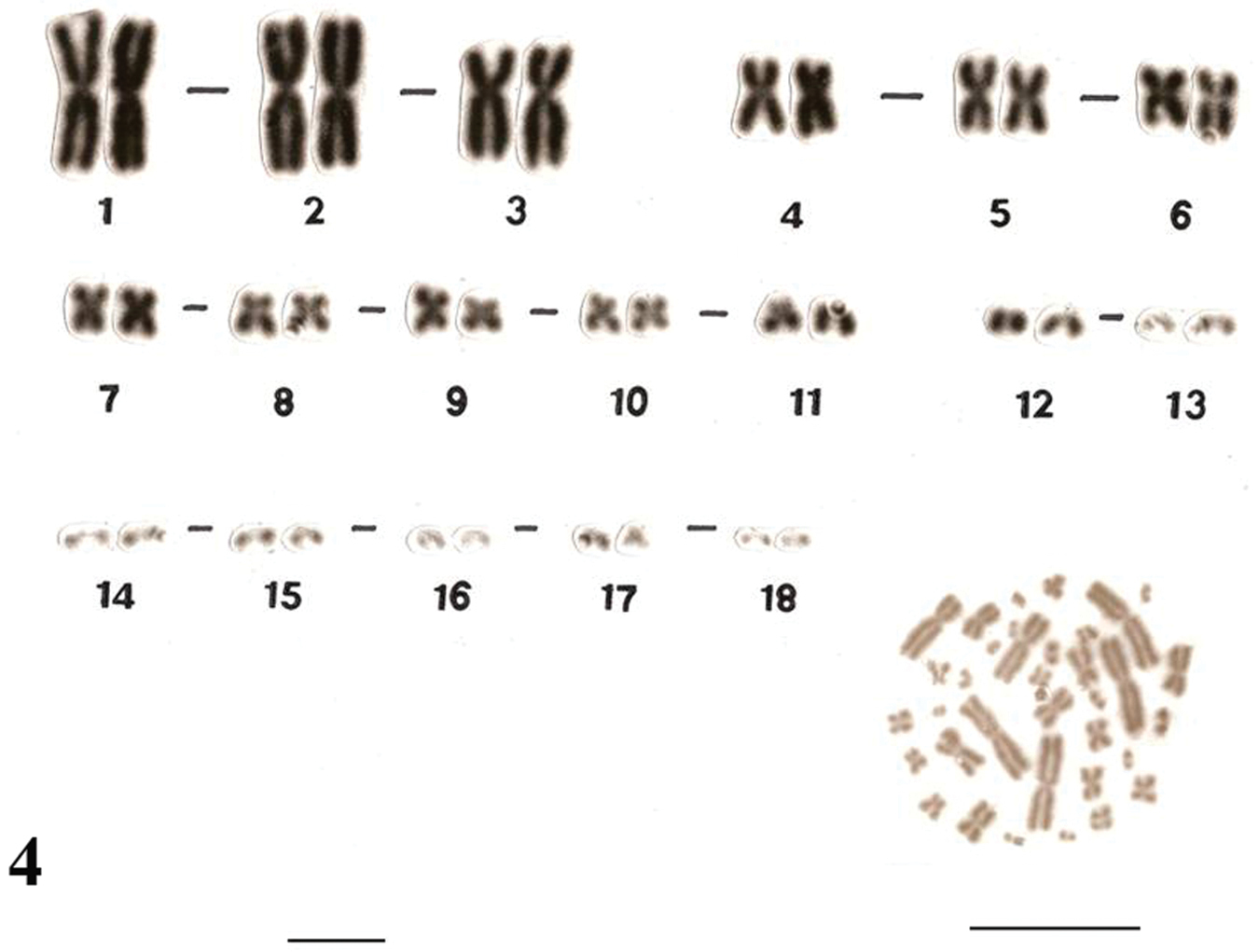
Similar karyotypes were obtained for the species Uraeotyphlus narayani (Fig. 3) and Uraeotyphlus menoni (Fig. 4).
No detectable sex chromosome pair was observed in either sex in the metaphase chromosomal complement and karyotypes of Uraeotyphlus oxyurus, Uraeotyphlus interruptus, Uraeotyphlus narayani and Uraeotyphlus menoni.
Giemsa stained male karyotype and female metaphase complement of Uraeotyphlus oxyurus. Bar = 10 µm.
Giemsa stained male karyotype and female metaphase complement of Uraeotyphlus interruptus. Bar = 10 µm.
Giemsa stained male karyotype and female metaphase complement of Uraeotyphlus narayani. Bar = 10 µm.
Giemsa stained male karyotype and female metaphase complement of Uraeotyphlus menoni. Bar = 10 µm.
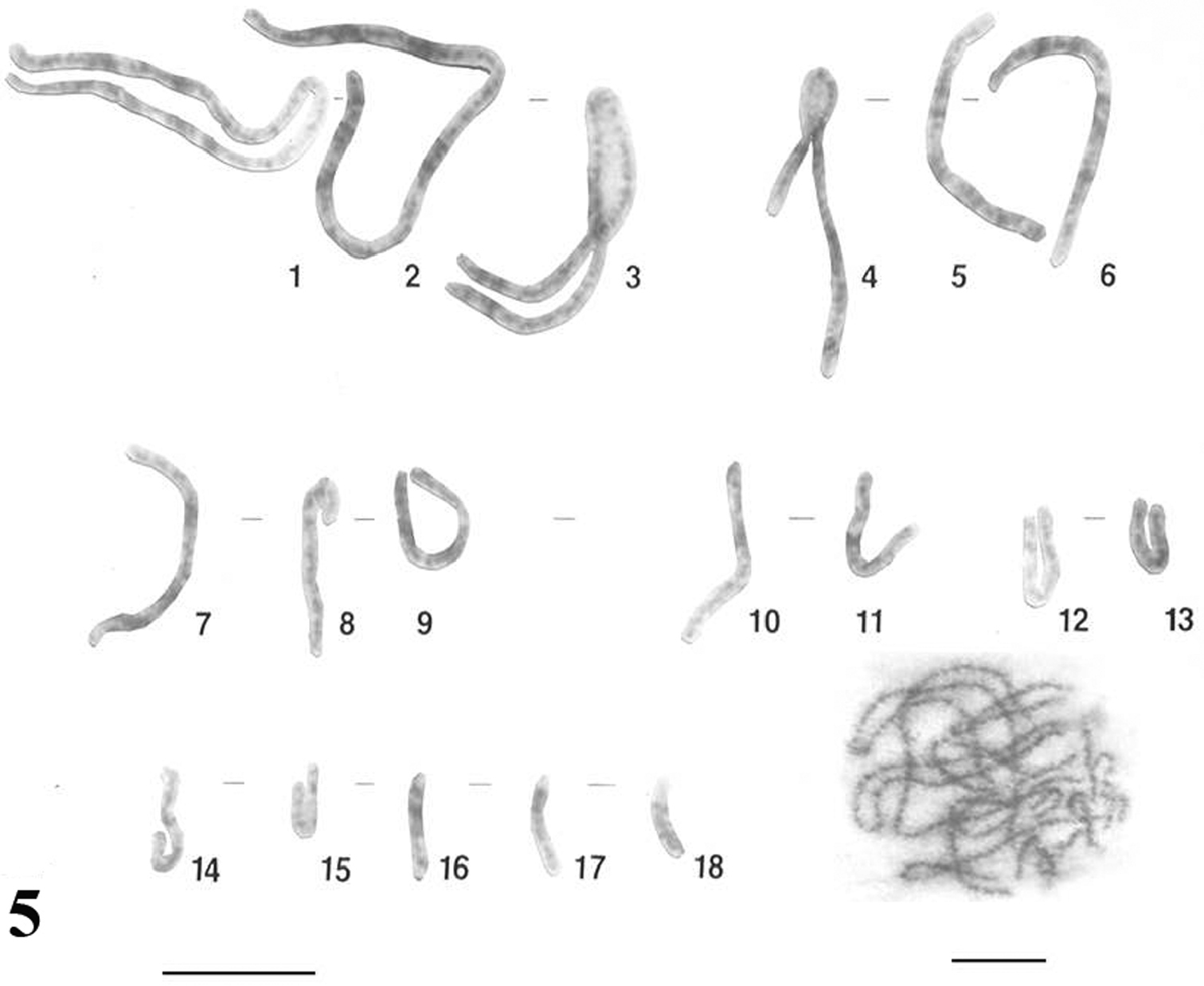
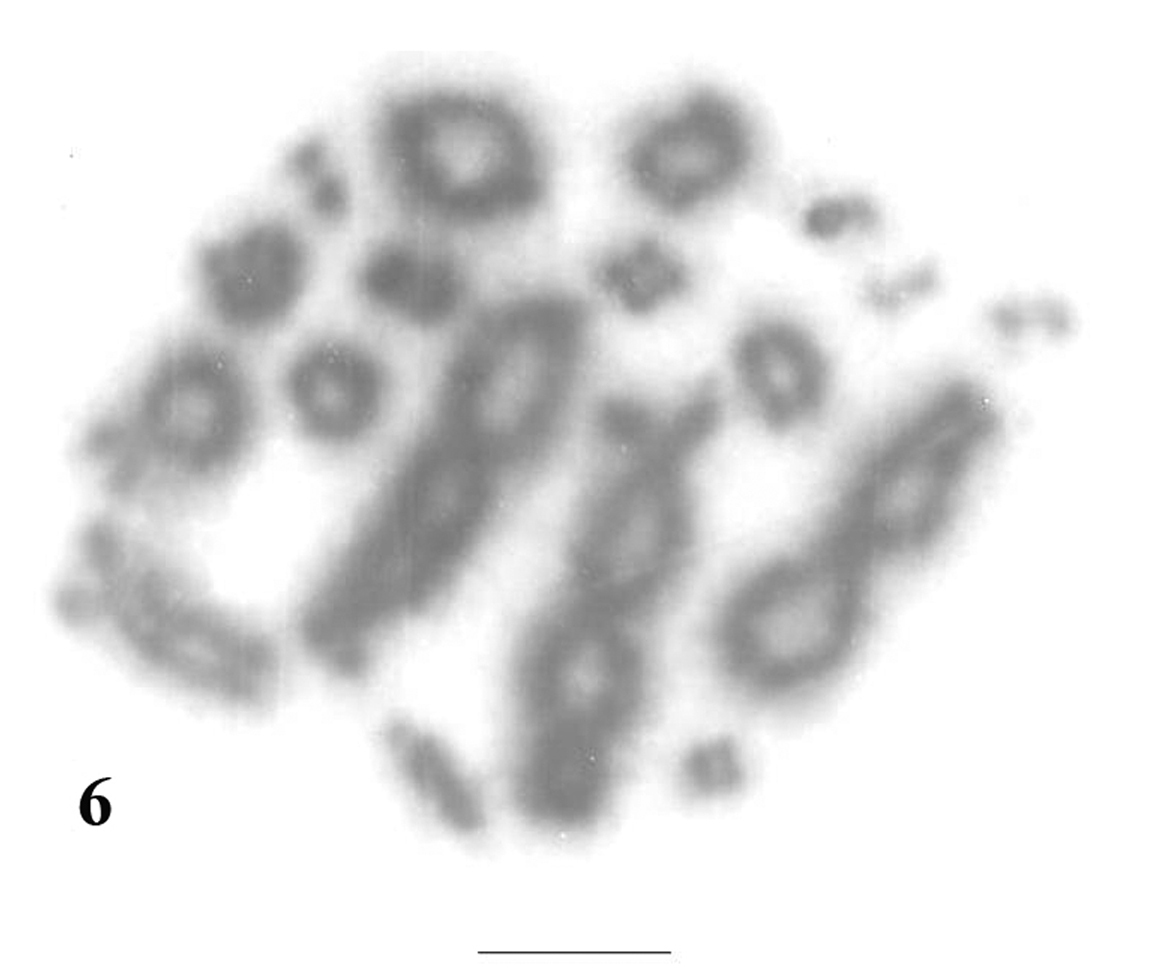
The meiotic chromosomes prepared from male individuals of Uraeotyphlus oxyurus revealed a good number of pachytene (Fig. 5), diplotene (Fig. 6), diakinetic and second meiotic metaphase configurations.
Pachytene chromosome karyotype constructed as per the somatic metaphase chromosome karyotype revealed eighteen pachytene bivalents corresponding the eighteen pairs of somatic chromosomes.
The diplotene complement allowed the counting of eighteen individually identifiable bivalents. In each diplotene complement, the longer ones carried 4-6 chiasmata, whereas the smaller acrocentrics consisting of at least one chiasma.
Similar results were obtained for the other three species, Uraeotyphlus interruptus, Uraeotyphlus narayani and Uraeotyphlus menoni.
Pachytene karyotype and complement of Uraeotyphlus oxyurus. Bar = 10 µm.
Diplotene complement of Uraeotyphlus oxyurus. Bar = 10 µm.
The Uraeotyphlus oxyurus karyotype is characterized by discernible but faintly stained centromeric C-bands in the metacentric and submetacentric chromosomes, while the acrocentrics have very prominent C-bands at the centromeric region cumulatively highlighting both the centromeric regions and the proximal portions of each short arms of each chromosome (Fig. 7). In comparison with the C-staining characteristics of Uraeotyphlus oxyurus, typical C-bands were observed in the other three species karyotypes.
C-stained male karyotype and female metaphase complement of Uraeotyphlus oxyurus. Bar = 10 µm.
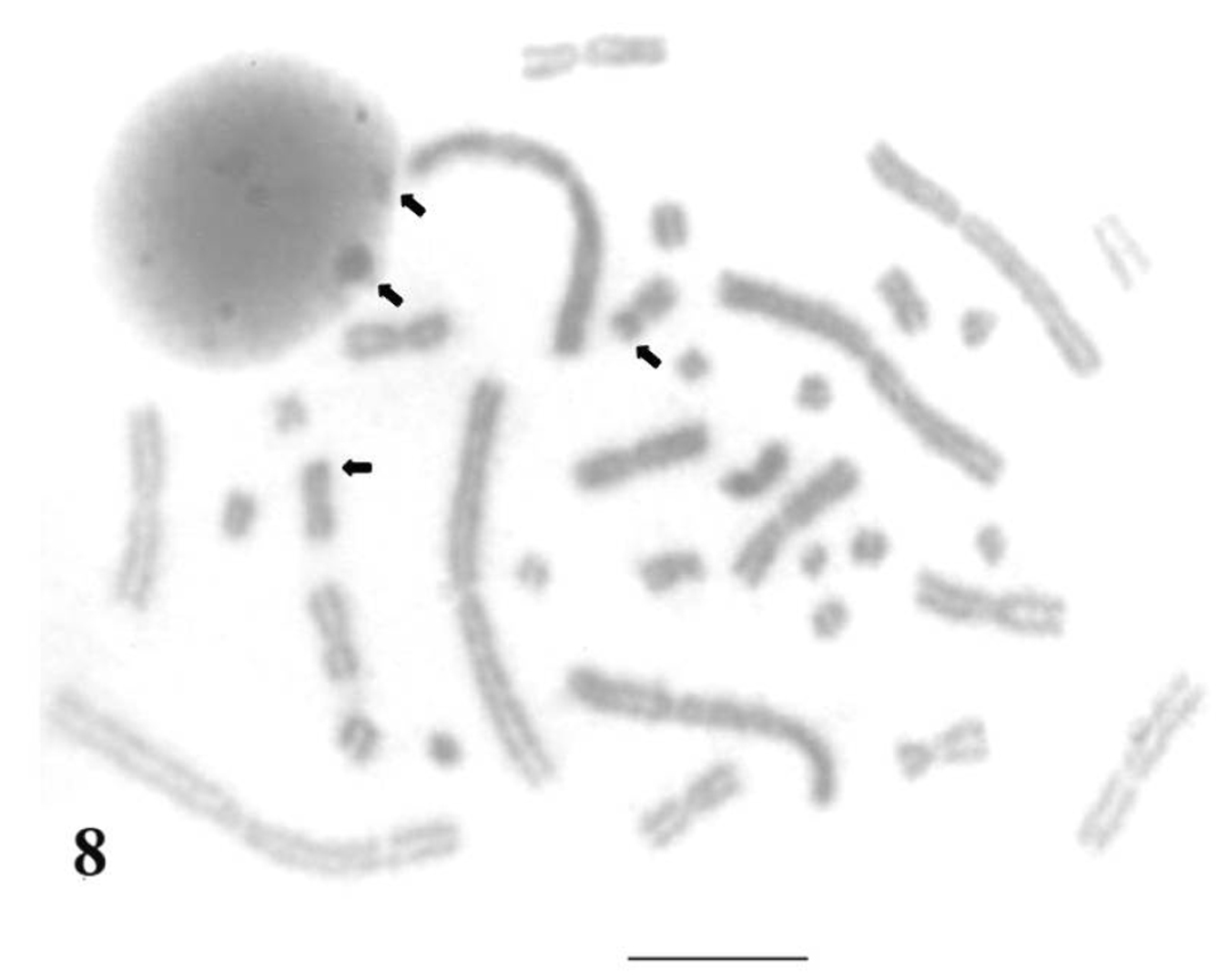
Silver nitrate staining showed that in Uraeotyphlus oxyurus, Nucleolus Organizer Regions (NORs) are confined to chrormosomal pair 9 in the complement but were not consistently demonstrated in each and every cell. Whereas, interphase nuclei demonstrated 1 to 2 (or sometimes 3) numbers of silver nitrate (2–3) aggregates (Fig. 8). This situation is perhaps an indication of limited proportionality of rDNA sites that could not be elicited cytologically and those of transcriptionally silent NORs did not form any discrete Ag-NORs sites at metaphase chromosomes.
Similar kind of results could be drawn for the other three species, Uraeotyphlus interruptus, Uraeotyphlus narayani and Uraeotyphlus menoni.
Silver-stained interphase nucleus and mitotic metaphase complement of Uraeotyphlus oxyurus. Bar = 10 µm.
Emphasizing on systematics of those Indian endemic uraeotyphlids, the morphological attributes as elicited by
In the present study, the karyotype carrying a diploid number of 36 (2n=36) chromosomes was found consistently identical in each of the four species belonging to Uraeotyphlus oxyurus species complex. The cytogenetic data converged on chromosome morphology indicate that the genus Uraeotyphlus is relatively well conserved with most chromosomal pairs classified as meta- and submetacentrics in size and shape. This observation, especially of all the four oxyurus type karyotypes having a homologous situation reveals a closer phylogenetic relationship. This, in turn, supports consideration for a monophyletic origin.
The same situation cannot be considered as applying to those species belonging to Uraeotyphlus malabaricus group. Until now, none of these species karyotypes were known except a publication citing a variable karyotype for the species belonging to the genus, Uraeotyphlus, Uraeotyphlus gansi.
This karyolgical description lends support to
The four species karyotypes (2n=36) may be considered as derived ones from that of Uraeotyphlus gansi karyotype (2n=42), which serves as a modal karyotype for the Uraeotyphlus malabaricus group species which could be considered a basal one among uraeotyphlids. Besides, based on the pronounced chromosomal homogeneity among the said species groups’ karyotypic specificities, this may be considered as of cytogenetic importance while placing emphasis on species differentiation within the genus. Uraeotyphlid chromosome complements belonging to family Ichthyophiidae seem to present bimodal diploid numbers: 2n=42 chromosomes in the basal type Uraeotyphlus malabaricus group and 2n=36 chromosomes in the case of derived species belonging to Uraeotyphlus oxyurus species complex.
Presently, chromosomal data on ichthyophiid taxa indicate that they are all well-conserved among the species whose karyotypes are known, bearing identical chromosomal set in each case (
During the course of drawing karyological relationships prevailing between the two taxa (for e.g., ichthyophiids and uraeotyphlids) it is possible to infer that most submeta and metacentrics are conserved to a maximum extent and only acrocentrics mayhave paved the way for chromosome speciation events.
There are instances that exhibit very little variation in their karyological features. Marked karyological homogeneity seems prevalent in many taxa of cyprinid fishes (
Any attempt on comparison of karyotypes from the present study to that of other representative karyotypes for Uraeotyphlus malabaricus type (although only one species karyotype is available) makes clear the occurrence of a succession of chromosomal rearrangements, mainly through pericentric inversion and / or fusion. This appears necessary in order to create the karyotype found in described karyotype from those of ancestral ones. This sequence of events could account for the appearance of reduction in basic chromosome numbers from 2n=42 chromosome to 2n=36 chromosomes (
The C-banding profile and NOR localization seems to be a homologous feature in oxyurus uraeotyphlids. C-banding pattern was found identical in the four species of oxyurus group within the genus. The variation illustrated by the two species group (viz., oxyurus and malabaricus) in which there is an enormous interspecific variation was evident in the distribution and amount of heterochromatin (
Based on certain molecular analysis, it is possible to ascribe that the basal Uraeotyphlus malabaricus group is closely aligned in its affinity to primitive ichthyophid lineages. However, in order to expend linealogical connections between these two broader groups,
Phylogeny of the Indian endemic genus Uraeotyphlus is still poorly known. But a combined approach based on morphology, biochemistry, molecular biology and cytogenetics might help to resolve a revised classification of Ichthyophiidae and thus to understand better of their phylogenetic relationships with other caecilians. Towards that effect,
While exploring probable phylogenetic relationships and rapport, it also seems possible to infer that karyological data fall in line with those of recently assimilated molecular analyses (
In conclusion, the cytogenetic study based on conventional Giemsa staining including C- and NOR bandings, upon four oxyurus group species of Uraeotyphlus taxa, indicate that they are more similar in their karyotypic profile (if not identical) which might form a monophyletic group. In the light of extensive chromosomal homology incurred, that does not preclude minor karyotypic differences necessitating in the use of other banding techniques for the improvement of karyological characterization.
We wish to thank Rashmi R. Nair, Suresh (Kottayam); Adeeb, Anoop, Amjesh, Abilash, Sona, Mahesh (Thiruvananthapuram); Sharon, Rajan, Jom George (Trichur); Raju, Santosh, Manoj (Goolikadavu); Priya Biju, Jose, Raju (Gudalur); Jaseer (Cochin); Vijaykumar, Gangadhar (Koodam village, Payyanur); Ramaswamy, Muniswamy Reddy, Joseph, Harry (Bangalore) for their help during specimen collection and to their families for the hospitality during our collection trips.