(C) 2013 Irina Bakloushinskaya. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
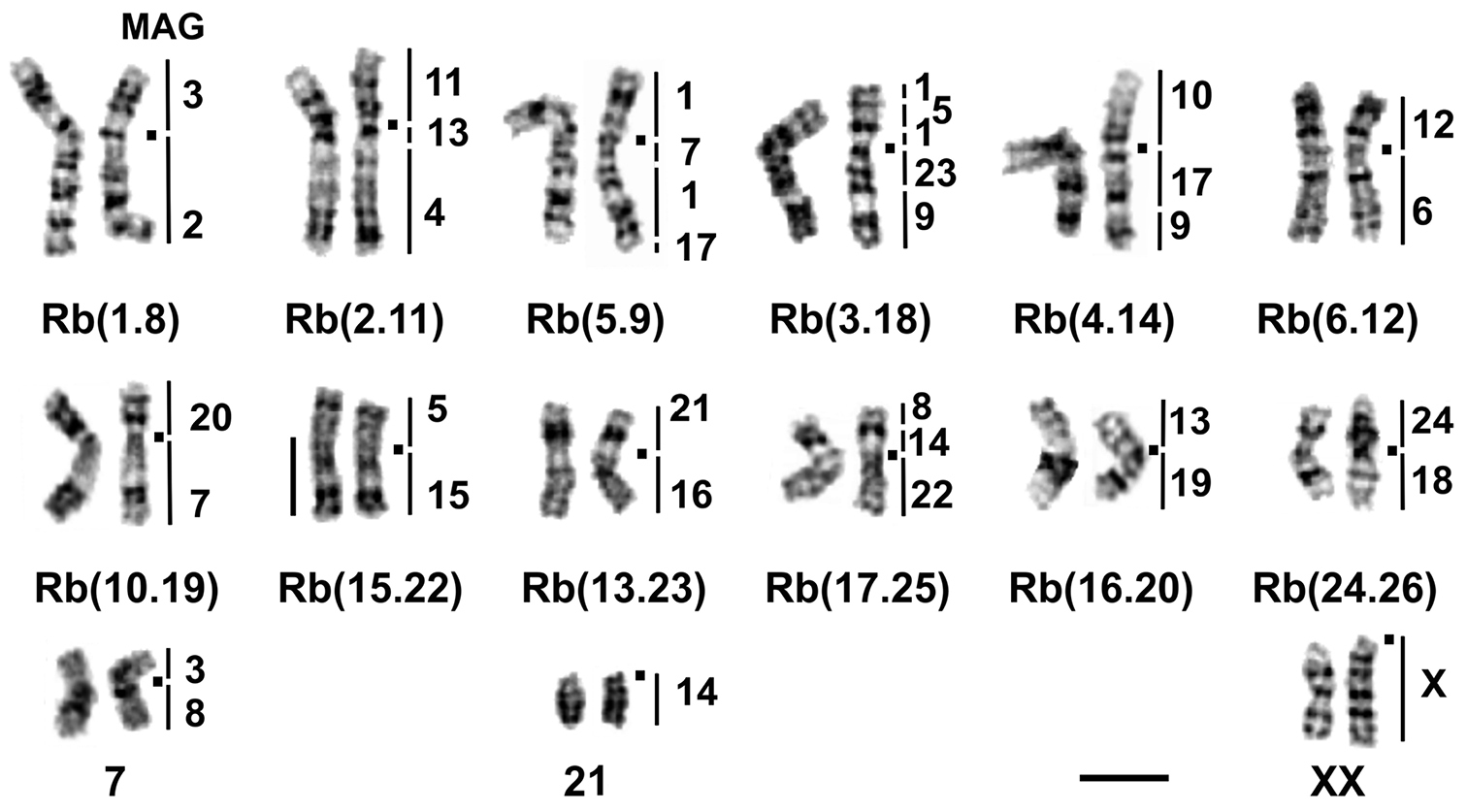
The subterranean mole vole, Ellobius tancrei, with aspecific variability in autosomes (2n = 31–54) and unusual sex chromosomes (XX in males and females), represents an amazing model for studying the role of chromosome changes in speciation. New materials from the upper reaches of the Surkhob River in the Pamiro-Alay mountains resulted in the discovery of a new form with 2n = 30. The application of Zoo-FISH and G-banding methods allowed the detection of 13 pairs of autosomes as Robertsonian metacentrics originated after fusions of acrocentrics of an assumed ancestral karyotype of Ellobius tancrei with 2n = 54. The sex chromosomes (XX, in both sexes) and one pair of acrocentric autosomes are the only acrocentrics in this karyotype, and the set with 2n = 30 possesses the lowest possible chromosome number among populations of Ellobius tancrei.
speciation, mole voles, Ellobius, Robertsonian rearrangements, chromosome painting, Cricetidae
Naturally occurring chromosome variability is essential for understanding the disputed role of chromosome changes in speciation (
Thorough analysis based on G-banding revealed a complicated structure of chromosome variability in the Surkhob River valley (Pamiro-Alay), where forms with the same chromosome numbers have different sets of Rb metacentrics. It was concluded that the variability was produced by hybridisation, as well as chain fusions (
Five animals (two females and three males) from two colonies were captured by live traps (
Chromosomes from bone marrow (
Images were captured using VideoTesT-FISH 2.0. and VideoTesT-Karyo 3.1. (VideoTesT) or Case Data Manager 6.0 (Applied Spectral Imaging Inc., ASI) software with either a ProgRes CCD (Jenoptik) or ASI CCD camera, respectively, mounted on an Axioskop 2 plus (Zeiss) microscope with filter sets for DAPI, FITC, and rhodamine. Hybridisation signals were assigned to specific chromosome regions defined by GTG-banding patterns previously photographed and captured with the CCD camera. All images were processed using PaintShop Photo Pro X2 (Corel).
We analysed the structure of karyotypes obtained by direct methods from bone marrow and from cultures. It is known that spontaneous chromosome aberrations may appear in cell cultures (
G-banded karyotype of Ellobius tancrei, 2n = 30 (25618 ♂). The chromosome nomenclature follows
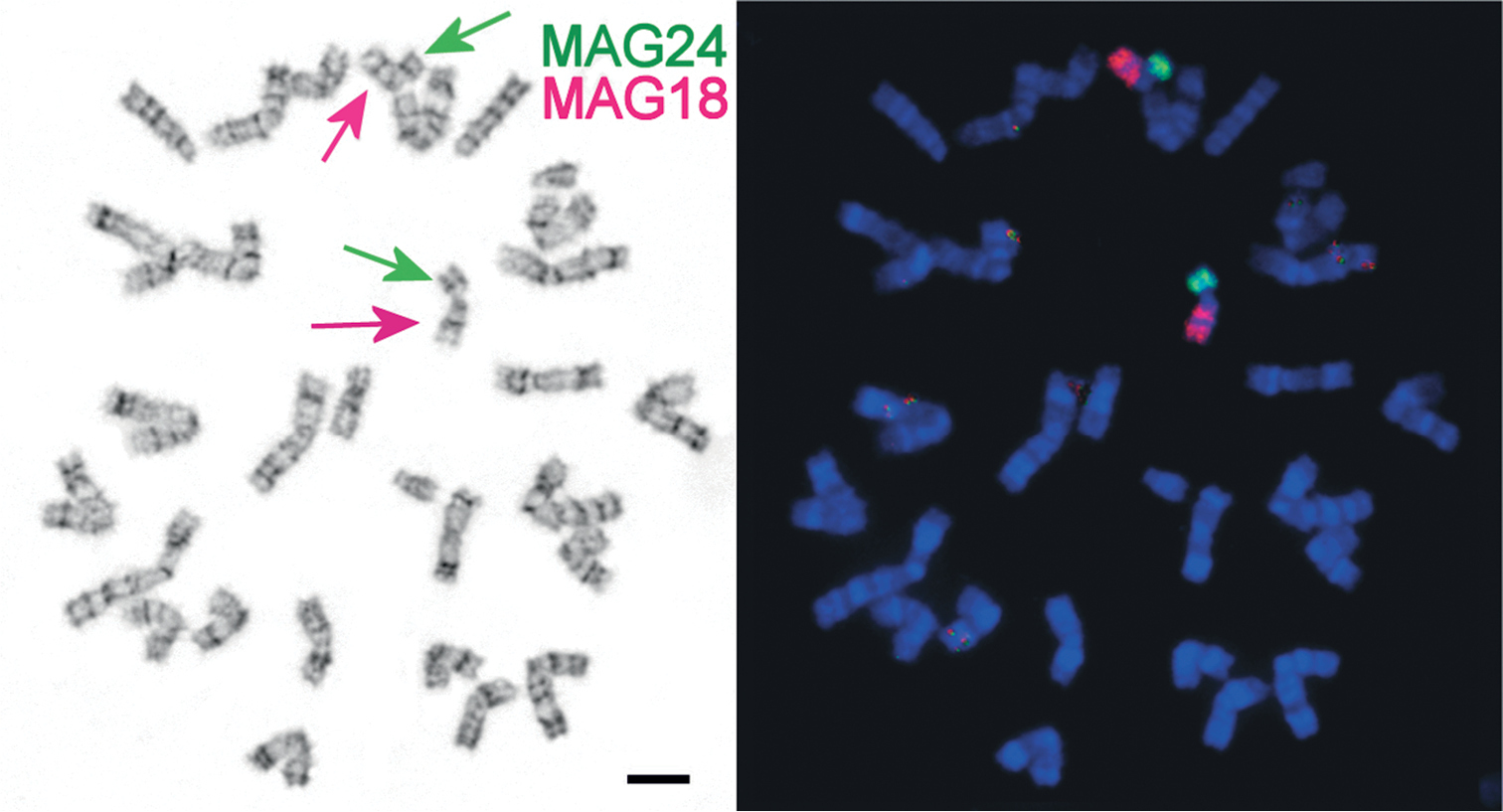
a G-banded Ellobius tancrei, 2n = 30 (25601 ♀) metaphase spread. b the same spread, fluorescent in situ hybridisation (Zoo-FISH) of Microtus agrestis (MAG) chromosome 18 (red) and 24 (green) on Rb metacentrics (red and green arrows). Bar = 10 μm.
The analysis of the spatio-temporal variation in the structure of a chromosomal polymorphism zone in Ellobius tancrei with an interval of 25 years showed that the distribution of chromosomal forms within the area has not changed during this period, except for a small range expansion of a low-chromosomal form (2n = 32) at the western boundary (
Applying the Zoo-FISH method allows the detection of the homology of translocations, which are only estimated by G-banding. The new karyotype with 2n = 30 contains three Rb translocations (metacentrics 2, 3, 4, Fig. 1) that are homologous to fusions recently described for the 2n = 48 populations inhabiting the northern bank of the Surkhob River, approximately 100 km to the west. Based on these data, we suggest a common origin for these populations. Independent origin may be suggested for a population from the southern bank of the Surkhob River (2n = 50), which shares monobrachial homology with the 2n = 48 form. Furthermore, a partial homology was revealed by Zoo-FISH in spite of their similar G-banding picture (
This study was funded in part by research grants from the Russian Foundation for Basic Research and Programs of the Russian Academy of Sciences (MCB and Dynamics and Conservation of Genofonds). We thank the Director of the Institute of Zoology and Parasitology of the Academy of Sciences of the Tadjik Republik, Dr. A.S. Saidov, for help in organizing the field research.