(C) 2013 Chirino Mónica Gabriela. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Male meiosis behaviour and heterochromatin characterization of three big water bug species were studied. Belostoma dentatum (Mayr, 1863), Belostoma elongatum Montandon, 1908 and Belostoma gestroi Montandon, 1903 possess 2n = 26 + X1X2Y (male). In these species, male meiosis is similar to that previously observed in Belostoma Latreille, 1807. In general, autosomal bivalents show a single chiasma terminally located and divide reductionally at anaphase I. On the other hand, sex chromosomes are achiasmatic, behave as univalents and segregate their chromatids equationally at anaphase I. The analysis of heterochromatin distribution and composition revealed a C-positive block at the terminal region of all autosomes in Belostoma dentatum, a C-positive block at the terminal region and C-positive interstitial dots on all autosomes in Belostoma elongatum, and a little C-positive band at the terminal region of autosomes in Belostoma gestroi. A C-positive band on one bivalent was DAPI negative/CMA3 positive in the three species. The CMA3-bright band, enriched in GC base pairs, was coincident with a NOR detected by FISH. The results obtained support the hypothesis that all species of Belostoma with multiple sex chromosome systems preserve NORs in autosomal bivalents. The karyotype analyses allow the cytogenetic characterization and identification of these species belonging to a difficult taxonomic group. Besides, the cytogenetic characterization will be useful in discussions about evolutionary trends of the genome organization and karyotype evolution in this genus.
Heteroptera, holokinetic chromosomes, karyotype evolution mechanisms, multiple sex chromosomes, rDNA-FISH
Belostomatidae include some of the largest heteropteran species, which are general predators that play an important role as biological agents in aquatic environments (
All species of Belostoma analyzed possess holokinetic chromosomes, i.e. chromosomes without a primary constriction and therefore without a localized centromere. Autosomal bivalents are synaptic and chiasmatic, whereas sex chromosomes are asynaptic and achiasmatic, and behave as univalents in first male meiotic division. However, at metaphase II sex chromosomes associate end-to-end through the so called touch-and-go pairing, forming a pseudo-bivalent or pseudo-multivalent. In the first meiotic division, autosomal bivalents segregate reductionally while sex chromosomes divide equationally (
Most hypotheses on karyotype evolution in Heteroptera include both autosomal and sex chromosome fusions and fragmentations (
The aim of this study was to perform a detailed comparison of male meiosis behaviour and examine the structure of the holokinetic chromosomes by means of C- and fluorescent bandings, and fluorescent in situ hybridization (FISH) with 18S rDNA probes in Belostoma dentatum (Mayr, 1863), Belostoma elongatum Montandon, 1908 and Belostoma gestroi Montandon, 1903. The female complement and the male meiosis of Belostoma elongatum and Belostoma gestroi are described for the first time. These results allowed us to distinguish morphologically similar species and, also, led us to propose a scenario of karyotype evolution in the genus Belostoma.
Diploid chromosome number, chromosome bandings and nucleolar organizer region (NOR) detected by FISH in South American Belostoma species. *A: autosomal bivalent, **X, Y: sex chromosomes
| Species | 2n (male) | C bands | DAPI/CMA3 bands | rDNA by FISH | References |
| Belostoma bergi (Montandon), 1899 | 26 + X1X2Y | no | no | -- | Papeschi and Bressa 2004 |
| Belostoma bifoveolatum Spinola, 1852 | 26 + X1X2Y | yes | yes | -- | Papeschi 1991, |
| Belostoma candidulum Montandon, 1903 | 14 + XY | yes | yes | -- |
|
| Belostoma cummingsi De Carlo, 1935 | 26 + X1X2Y | no | no | -- |
|
| Belostoma dentatum (Mayr, 1863) | 26 + X1X2Y | yes | yes | A* |
|
| Belostoma dilatatum (Dufour, 1863) | 26 + X1X2Y | yes | no | -- |
|
| 26 + X1X2 X3Y | yes | yes | -- |
|
|
| Belostoma discretum Montandon, 1903 | 26 + X1X2Y | yes | yes | -- |
|
| Belostoma elegans (Mayr, 1871) | 26 + X1X2Y | yes | yes | A* |
|
|
|
|||||
| Belostoma elongatum Montandon, 1908 | 26 + X1X2Y | yes | yes | A* |
|
| Belostoma gestroi Montandon, 1903 | 26 + X1X2Y | yes | yes | A* |
|
| Belostoma martini (Montandon, 1899) | 26 + X1X2Y | yes | no | -- | Papeschi 1991 |
| Belostoma micantulum (Stål, 1860) | 14 + XY | yes | yes | X, Y* |
|
| Belostoma orbiculatum Estévez & Polhemus, 2001 | 14 + XY | yes | yes | -- |
|
| 14 + X1X2Y |
|
||||
| Belostoma oxyurum (Dufour, 1863) | 6 + XY | yes | yes | X, Y** |
|
| Belostoma plebejum (Stål, 1858) | 14 + XY | no | no | -- | |
| 13 + XY |
|
||||
| 14 + X1X2Y |
For meiotic analysis, adults and nymphs of Belostoma elongatum (9 males and 8 females) and Belostoma gestroi (4 males and 12 females) were collected from 1988 to 1990 in several fields from Buenos Aires, Santa Fe, Entre Ríos, Corrientes and Misiones provinces, all in Argentina (Table 2). For chromosome bandings and fluorescent in situ hybridization (FISH) technique, adults of Belostoma dentatum (3 males and 1 female), Belostoma elongatum (3 males) and Belostoma gestroi (2 males and 1 female) were collected from 2010 to 2011 in Corrientes province (Argentina) (Table 2). Collected adults were identified according to the keys provided by
Species, provenience, geographical coordinates, and number of adults’ collected and examined of Belostoma for chromosomal analyses discriminated by gender.
| Species | Chromosomal analyses | Localities from Argentina | Coordinates | N° of individuals |
| Belostoma dentatum | C- and DAPI-CMA3 bandings | San Pedro, Buenos Aires | 33°40'33"S, 59°39'47"W | 3 males |
| FISH technique | Corrientes, Corrientes | 27°28'16"S, 58°50'22"W | 1 female | |
| Belostoma elongatum | Chromosome complement | Arroyo Cuay Grande, Corrientes | 28°28'16"S, 58°50'22"W | 1 female |
| Male meiotic behaviour | Lagos de Stieler, Misiones | 26°34'2"S, 54°45'57"W | 1 male | |
| Valle Hermoso, Misiones | 26°23'10"S, 54°27'58"W | 8 males, 7 females | ||
| C- and DAPI-CMA3 bandings FISH technique | Corrientes, Corrientes | 27°28'16"S, 58°50'22"W | 3 males | |
| Belostoma gestroi | Chromosome complement | Río San Pedro, Buenos Aires | 33°40'33"S, 59°39'47"W | 1 male |
| Male meiotic behaviour | Rincón Norte, Santa Fe | 31°36'4"S, 60°34'12"W | 3 males, 11 females | |
| Santa Rosa, Santa Fe | 31°26'00"S, 60°22'00"W | 1 female | ||
| C- and DAPI-CMA3 bandingsFISH technique | Corrientes, Corrientes | 27°28'16"S, 58°50'22"W | 2 males, 1 female |
The captured specimenswere brought alive to the laboratory and reared until their gonads were dissected out. For meiotic analysis, the adults and nymphs were fixed for 15–30 min in freshly prepared fixative (ethanol:glacial acetic acid, 3:1). Afterwards, gonads were dissected out and kept at 4° C in 70% ethanol. Slides were prepared by the squash technique in a drop of 2% iron-propionic haematoxylin following conventional procedures (
Heterochromatin content, distribution and nucleotide composition were analysed by means of C- and sequential fluorescent DAPI and CMA3 bandings. C-banding was performed according to
Unlabelled 18S ribosomal DNA (rDNA) probes were generated by polymerase chain reaction (PCR) using universal arthropod primers: forward 5´-CCTGAGAAACGGCTACCACATC-3´ and reverse 5´-GAGTCTCGTTCGTTATCGGA-3´ (Whiting 2002). Total genomic DNA of Dysdercus albofasciatus Berg, 1878, obtained by standard phenol-chloroform-isoamylalcohol extraction, was used as a template. PCR was done following the procedure described in
Data of C-positive heterochromatin percentage and the haploid DNA content in Belostoma dentatum, Belostoma elongatum and Belostoma gestroi are part of the results obtained by Papeschi in her
The total chromosome length measurements (TCL) were performed with Micro Measure for Windows, version 3.3. The TCL of all bivalents and sex chromosomes were performed in metaphase I. Differences in TCL among species were compared by using one-way analysis of variance (ANOVA), with Fisher adjusted a posterior contrast. Statistical analyses were done using Statview software (SAS Institute Inc., 1992-1998).
Preparations were observed in epifluorescence microscopes: Zeiss Laborlux (Carl Zeiss, Germany) equipped with an analogue camera and Leica DMLB equipped with a Leica DFC350 FX CCD camera and Leica IM50 software, version 4.0 (Leica Microsystems Imaging Solutions Ltd., Cambridge, UK). Photomicrographs from meiotic chromosome preparations were taken using Kodak colour Supra print film 400 ASA. Black-and-white images of chromosomes from C- and fluorescent bandings and FISH technique were recorded separately for each fluorescent dye with the CCD camera. Images were pseudo-coloured (light blue for DAPI, green for CMA3, and red for Cy3), and processed with an appropriate software.
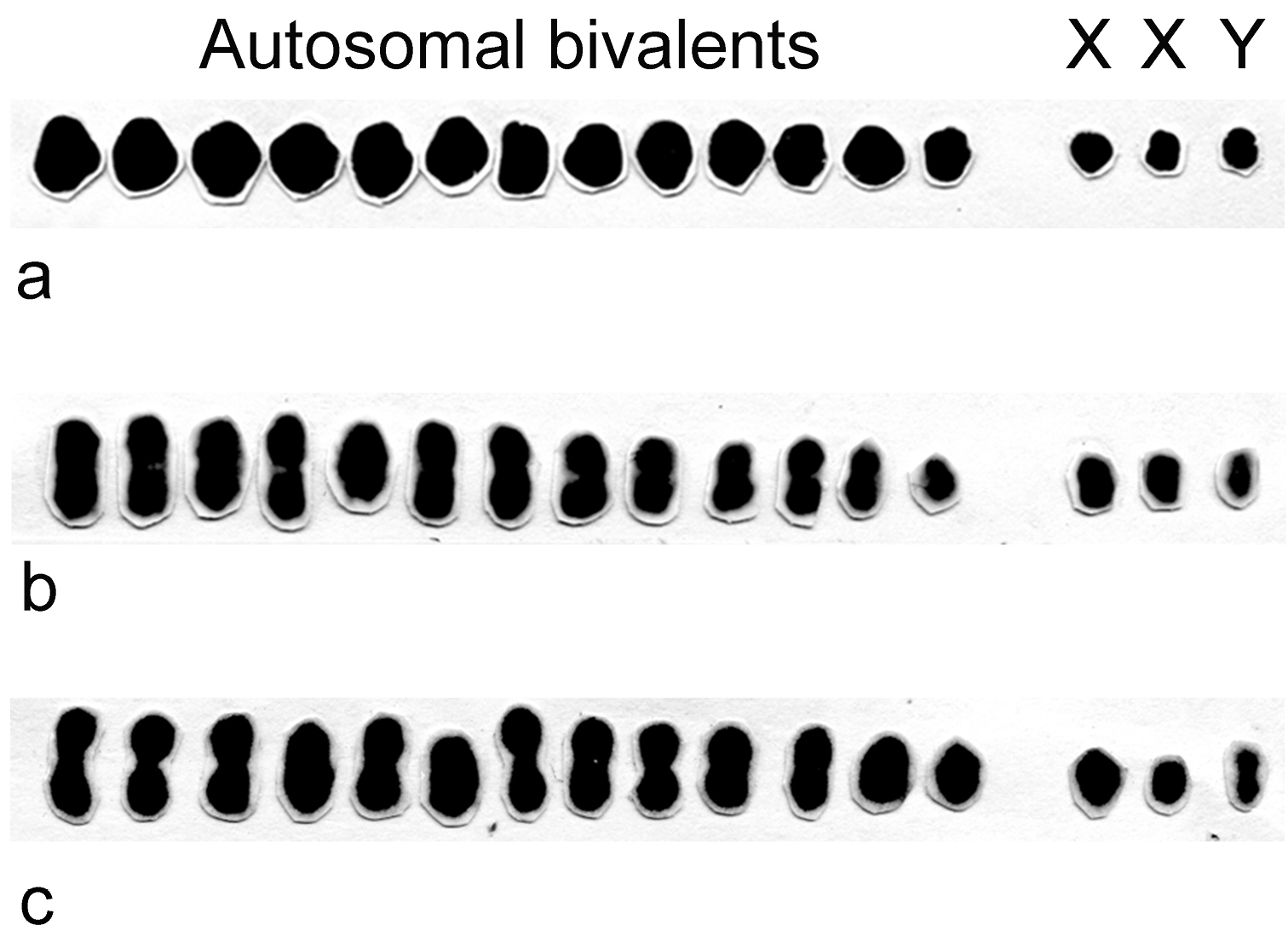
Male meiotic karyotypes based on metaphase I autosomal bivalents (II) and sex univalents of Belostoma dentatum, Belostoma elongatum and Belostoma gestroi show a male diploid chromosome number 2n = 13II + X1X2Y (Fig. 1). In the three species, the autosomes decrease gradually in size, both X chromosomes differ slightly in size and the Y chromosome is the smallest of the complement. The chromosome complement and male meiotic behaviour of Belostoma dentatum have already been described (
Male meiotic karyotypes of Belostoma dentatum (a), Belostoma elongatum (b) and Belostoma gestroi (c), 2n = 13II + X1X2Y, stained with 2% iron-propionic haematoxylin.
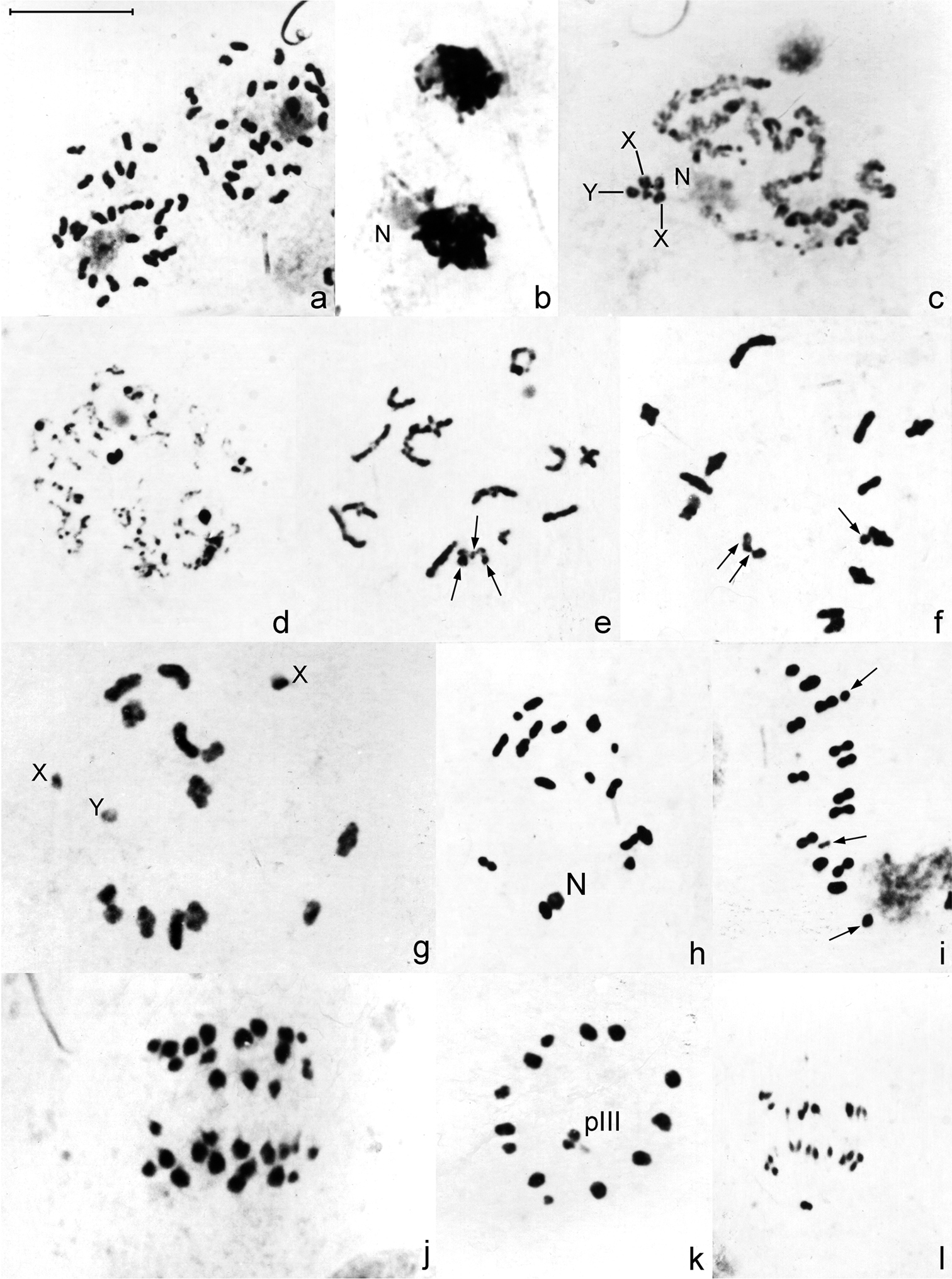
Analysis of spermatogonial prometaphase of Belostoma elongatum and Belostoma gestroi revealed a diploid number of 29 chromosomes; both karyotypes were as described by
Male meiosis in Belostoma elongatum (b, c, g, j, k) and Belostoma gestroi (a, d, e, f, h, i, l) stained with 2% iron-propionic haematoxylin. a Spermatogonial prometaphase b Synizesis c Pachytene, X and Y = sex chromosomes d Diffuse stage e–f Early diakinesis g–h Diakinesis i Metaphase I j Anaphase I k Metaphase II, Y sex chromosome is negatively heteropycnotic l Anaphase II. Arrows indicate sex chromosomes. pIII = pseudo-trivalent. N = nucleolus. Bar = 10 μm.
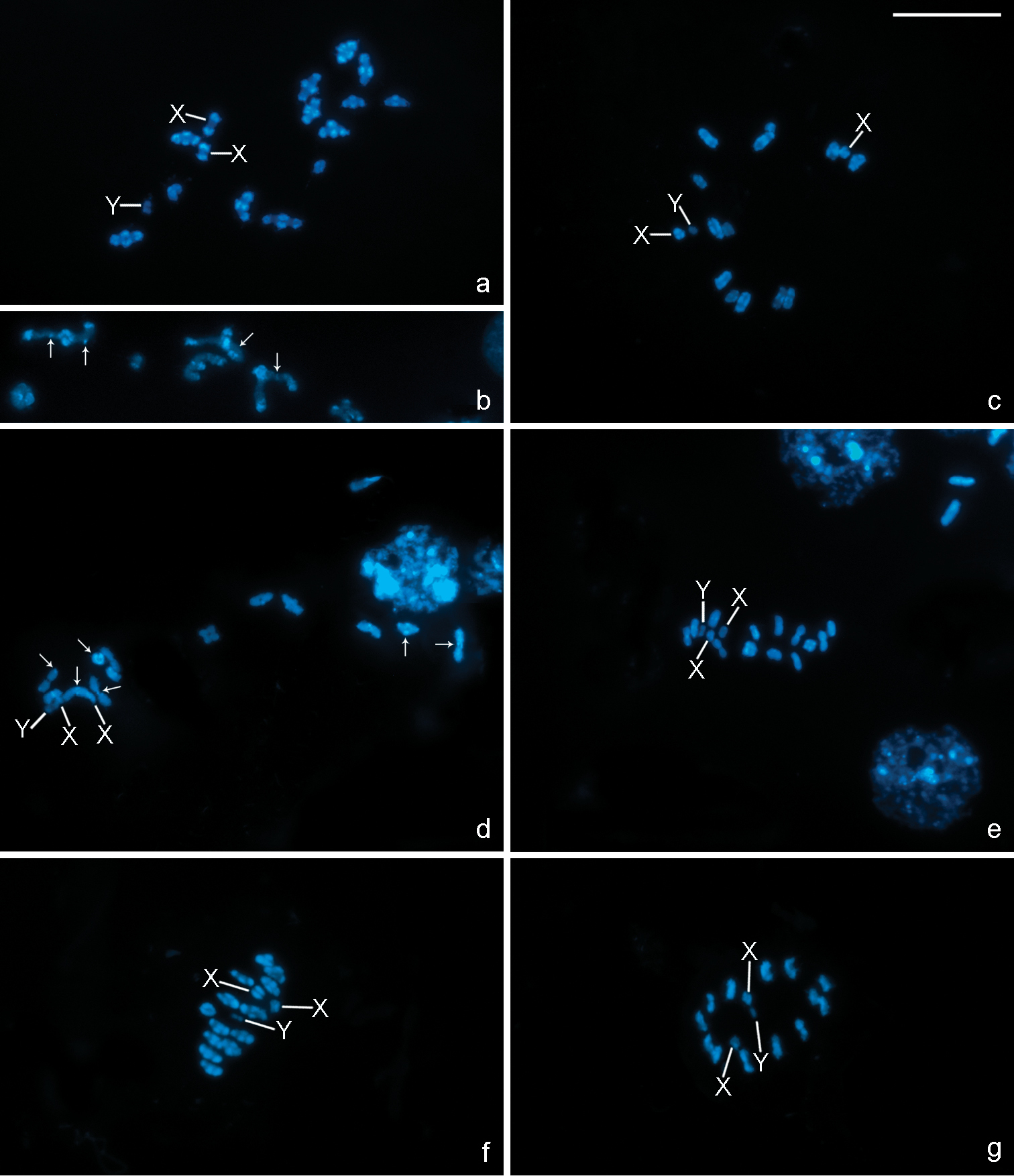
C-banding reveals differences in the amount and location of heterochromatin among the three species analysed. In Belostoma elongatum, very large C-positive blocks can be detected terminally on all bivalents from prophase I to metaphase I, and interstitial dots are also observed (Fig. 3a–c). In Belostoma gestroi, in contrast, C-positive bands are very small and are always located terminally (Fig. 3d, e). The results observed in Belostoma dentatum matched data previously described by Papeschi (1991) with C-positive bands terminally located in all bivalents (Fig. 3f, g). Furthermore, the two X chromosomes in the three species show terminally located bands, whereas the Y chromosome is C-negative (Fig. 3a, c–g).
All chromosomes stain homogenously with both fluorochromes on mitotic and meiotic metaphase cells in the three species, except for one of the medium-sized autosomal bivalents in Belostoma dentatum (Fig. 4a–c) and Belostoma elongatum (Fig. 4d–f), and one of the large-sized in Belostoma gestroi (Fig. 4g–i), which show a DAPI negative/CMA3 positive band at one terminal position.
C-banding in chromosomes of Belostoma elongatum (a–c), Belostoma gestroi (d, e) and Belostoma dentatum (f, g) stained with DAPI. a Diakinesis, conspicuous terminal C-positive blocks are observed in all autosomal bivalents and both X chromosomes b A detail of autosomal bivalents with interstitial C-positive dots (arrows) at early diakinesis c Late diakinesis d Diakinesis, small terminal C-positive bands in some autosomal bivalents (arrows) e Metaphase I f Late diakinesis, terminal C-positive bands in all autosomal bivalents and both X chromosomes g Metaphase II. a, c–g The Y chromosome is C-negative. X, Y = sex chromosomes. Bar = 10 μm.
DAPI (blue) and CMA3 (green) fluorescent banding in chromosomes of Belostoma dentatum (a–c), Belostoma elongatum (d–f) and Belostoma gestroi (g–i). a Oogonial metaphase (2n = 30 = 26 + X1X1X2X2) b Diakinesis c Metaphase II d Spermatogonial metaphase (2n = 29 = 26 + X1X2Y) e Diakinesis f Metaphase II g Spermatogonial metaphase (2n = 29 = 26 + X1X2Y) h Diakinesis i Metaphase II. Arrows indicate DAPI negative/CMA3 positive bands. Arrowheads show sex chromosomes (d, g). X, Y = sex chromosomes. pIII = pseudo-trivalent. Bar = 10 µm.
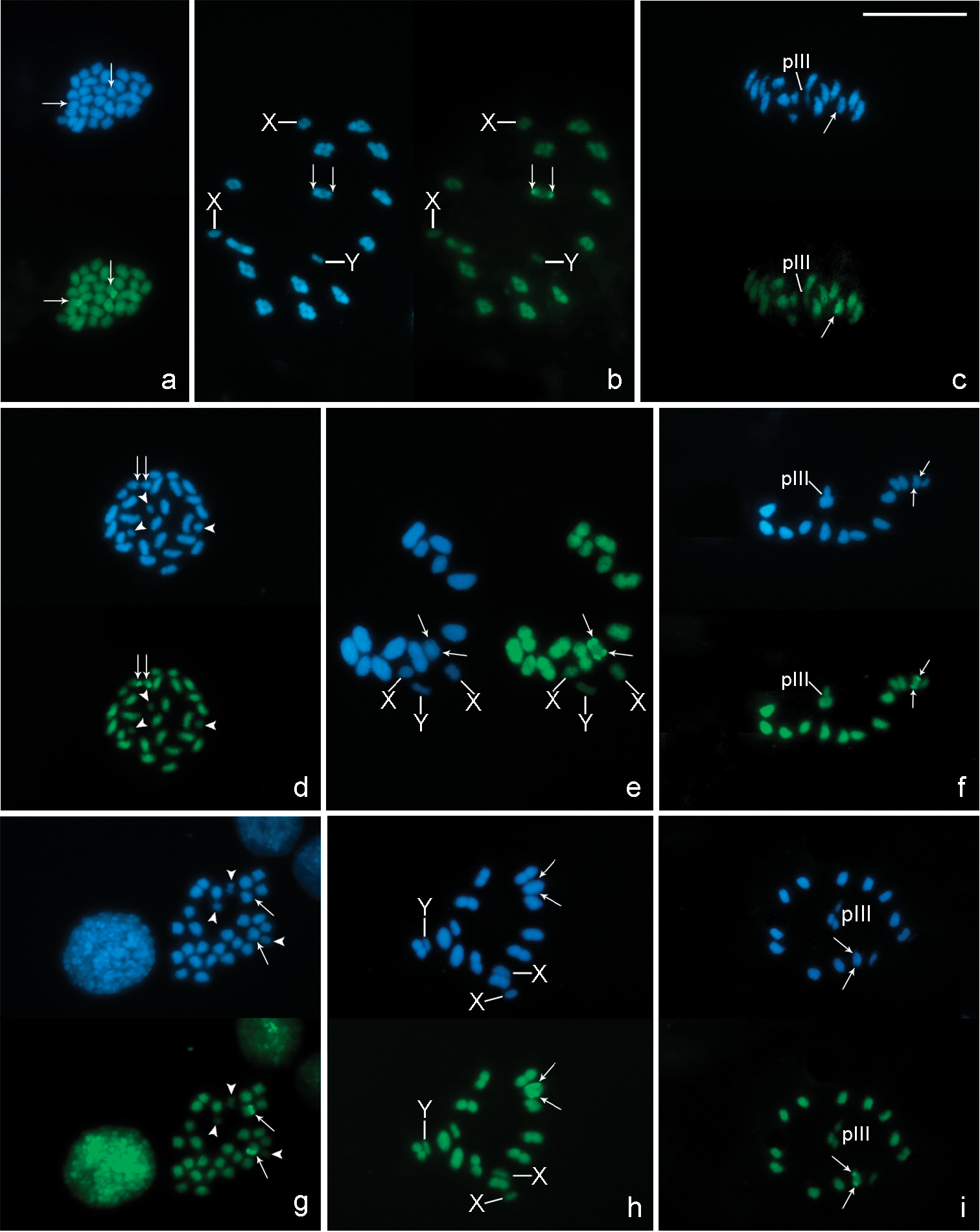
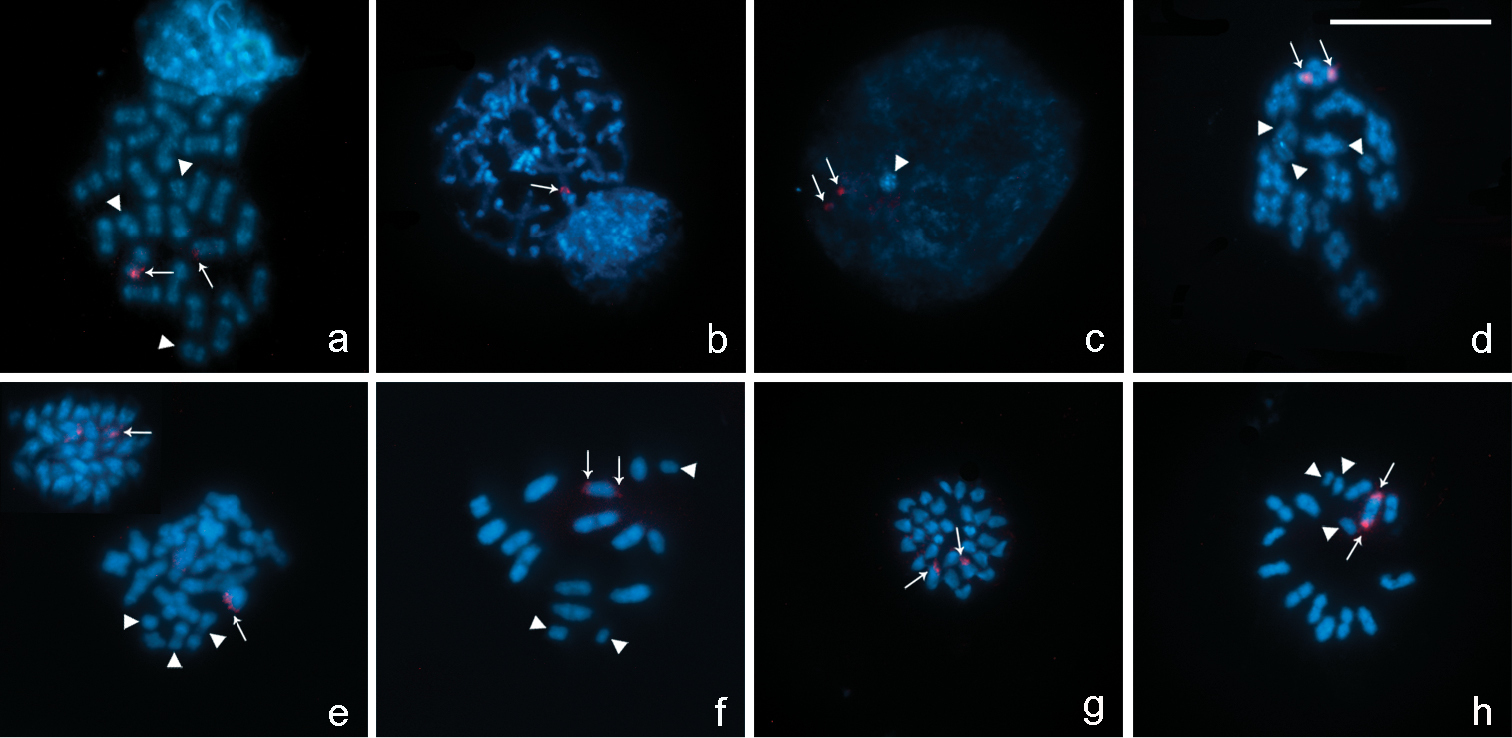
In chromosome preparations of Belostoma dentatum, FISH experiments with the 18S rDNA probe show a cluster of rDNA genes located at one end of two homologous chromosomes each (Fig. 5a). A single cluster of signals is observed in an autosomal bivalent at pachytene (Fig. 5b). During diffuse stage, hybridization signals are observed in the decondensed mass of autosomal chromatin, whereas the sex chromosomes remain condensed forming a conspicuous DAPI bright chromatin body without any signals (Fig. 5c). At diakinesis-metaphase I, one medium-sized autosomal bivalent show hybridization signals at both ends (Fig. 5d). In concordance with the results of Belostoma dentatum, in mitotic metaphases of Belostoma elongatum and Belostoma gestroi, hybridization signals are detected in two homologous autosomes (Fig. 5e, g). At diakinesis-metaphase I, a single cluster of rRNA genes is located at both ends of a medium-sized autosomal bivalent of Belostoma elongatum (Fig. 5e–f) and of a one large-sized of Belostoma gestroi (Fig. 5h).
Location of rDNA genes in chromosomes of Belostoma dentatum (a–d), Belostoma elongatum (e, f) and Belostoma gestroi (g, h) by FISH with 18S rDNA probes (red signals, arrows). Chromosomes were counterstained with DAPI (blue). a Spermatogonial anaphase (2n = 29 = 26 + X1X2Y) b Pachytene c Diffuse stage d Diakinesis e Spermatogonial metaphase and diakinesis f Metaphase I g Spermatogonial metaphase (2n = 29 = 26 + X1X2Y) h Diakinesis-Metaphase I. Arrowheads show sex chromosomes. Bar = 10 µm.
The Belostoma species analyzed here shared apparently similar karyotypes, since they possess the same chromosome complement (2n = 29 = 26 + X1X2Y, male), with chromosomes progressively decreasing in size. In Belostomatidae, this 2n is the modal diploid chromosome number and is present in 10 species of Belostoma (
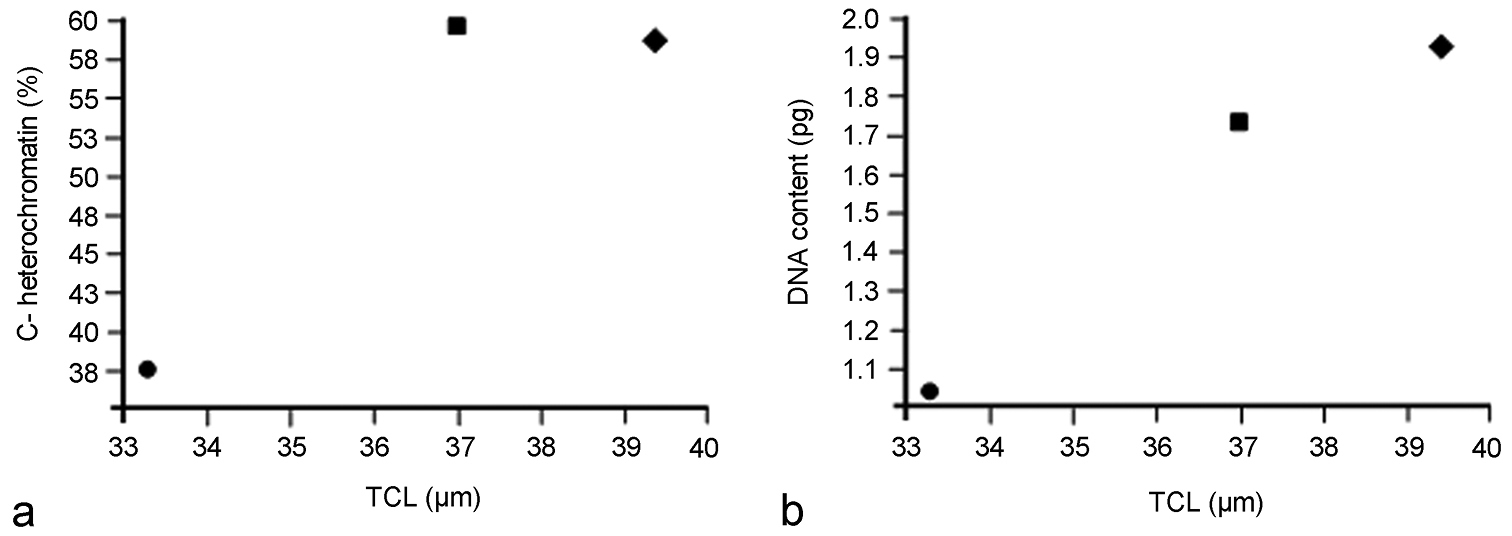
Heterochromatin characterization in the three species revealed differences in the amount, distribution and location of the constitutive heterochromatin in autosomes and both X chromosomes: i) terminal C-positive bands in Belostoma dentatum, ii) conspicuous C-positive bands at terminal and interstitial positions in Belostoma elongatum, and iii) very scarce C-positive bands terminally located in Belostoma gestroi. This variation in the constitutive heterochromatin of these three species could imply changes in the DNA content in the karyotype evolution in the genus, which could modify the size of the chromosome complement. In accordance with this suggestion, the analysis of TCL showed a significant variation among the three species, which means that certain chromosomal changes, must have taken place during their evolution.
In Heteroptera, the classical distribution pattern of C-positive heterochromatin is terminal in some or all chromosomes. Interstitial C-positive bands are described in a few species and some of them correspond to secondary constrictions and NORs. In concordance with these cytogenetic features, the C-banding pattern observed in Belostoma elongatum with respect to both terminal and interstitial C-positive regions agrees with most previous reports within Belostoma (
a Comparison between the total chromosome length (TCL) and the percentage of C-positive heterochromatin at diakinesis in Belostoma dentatum (58.54 ± 1.27 %; circle), Belostoma elongatum (59.47 ± 0.78 %; rectangle) and Belostoma gestroi (37.24 ± 1.50 %; diamond) b Comparison between the total chromosome length (TCL) and the haploid DNA content (pg) in Belostoma dentatum (1.93 ± 0.16 μm; circle), Belostoma elongatum (1.75 ± 0.05 μm; rectangle) and Belostoma gestroi (1.13 ± 0.13 μm; diamond). Data of percentage of C-positive heterochromatin and the haploid DNA content were obtained from Papeschi (1991,
The results with fluorescent banding indicate that all C-positive bands in the species analysed were not enriched with AT or CG base pairs, as all chromosomes were stained homogeneously with both DAPI and CMA3 fluorochromes, except for the C- positive band observed in the medium-sized autosomal bivalent of Belostoma dentatum and Belostoma elongatum and in one of the large-sized of Belostoma gestroi, which was DAPI negative/CMA3 positive. Therefore, the CMA3 bright band is enriched in GC base pairs and could represent an NOR (see below). The presence of a CMA3 bright band was also reported not only in other species of Belostoma (
In Belostomatidae, the location of NORs was previously analysed by FISH with 18S rDNA probe in Belostoma oxyurum (Dufour, 1863) (2n = 6 + XY, NOR in sex chromosomes), Belostoma micantulum (Stål, 1860) (2n = 14 + XY, NOR in sex chromosomes), Belostoma elegans (Mayr, 1871) (2n = 26 + X1X2Y, NOR in a pair of autosomes) (
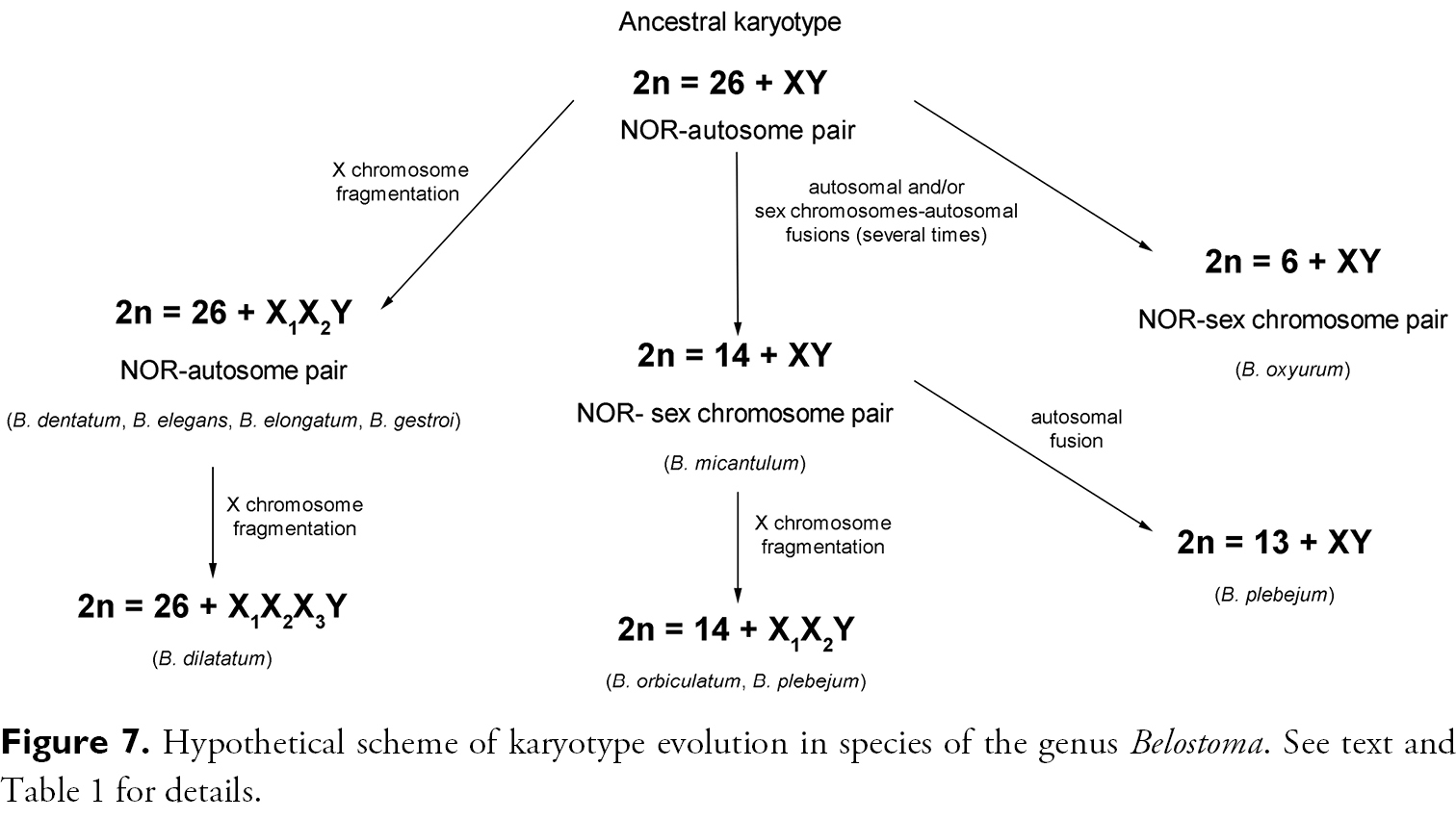
Previous cytogenetic data on worldwide Belostomatidae species allowed
Published data on karyotype evolution in species of this genus (
Conventional taxonomy of water bugs has focused almost entirely on adult specimens. There are relatively few publications on interspecific differences among the larvae, and fewer still concern South American species. The literature of the genus Belostoma includes much confusion because, in many cases, the species are very similar in coloration and appearance and only males or rarely only females can be identified (
Hypothetical scheme of karyotype evolution in species of the genus Belostoma. See text and Table 1 for details.
This work was funded by grants UBACyT W917 of University of Buenos Aires, PIP 0281 of Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), and PICT 2007-00635 of ANPCyT from Argentina. MG Chirino and MJ Bressa thank CONICET and ANPCyT. We wish to thank A. Bachmann and C Armúa de Reyes for taxonomic identification of the specimens included in the study.