(C) 2013 Victor Manuel Gomez-Rodriguez. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Gomez-Rodriguez VM, Rodriguez-Garay B, Palomino G, Martínez J, Barba-Gonzalez R (2013) Physical mapping of 5S and 18S ribosomal DNA in three species of Agave (Asparagales, Asparagaceae). Comparative Cytogenetics 7(3): 191–203. doi: 10.3897/CompCytogen.v7i3.5337
Agave Linnaeus, 1753 is endemic of America and is considered one of the most important crops in Mexico due to its key role in the country’s economy. Cytogenetic analysis was carried out in Agave tequilana Weber, 1902 ‘Azul’, Agave cupreata Trelease et Berger, 1915 and Agave angustifolia Haworth, 1812. The analysis showed that in all species the diploid chromosome number was 2n = 60, with bimodal karyotypes composed of five pairs of large chromosomes and 25 pairs of small chromosomes. Furthermore, different karyotypical formulae as well as a secondary constriction in a large chromosome pair were found in all species. Fluorescent in situ hybridization (FISH) was used for physical mapping of 5S and 18S ribosomal DNA (rDNA). All species analyzed showed that 5S rDNA was located in both arms of a small chromosome pair, while 18S rDNA was associated with the secondary constriction of a large chromosome pair. Data of FISH analysis provides new information about the position and number of rDNA loci and helps for detection of hybrids in breeding programs as well as evolutionary studies.
Agave, Fluorescent In Situ Hybridization, Ribosomal DNA, Karyotype, Physical mapping
Agave Linnaeus, 1753 is a genus of the monocotyledonous family Asparagaceae, belonging to the subfamily Agavoideae (
Fluorescent in situ hybridization (FISH) is a very useful technique in plant cytogenetics for the physical mapping of multigene families (
The aim of this work was to identify the number and chromosomal location of rDNA sites in three different species of the genus Agave including Agave tequilana Weber, 1902 ‘Azul’, Agave angustifolia Haworth, 1812 ‘Lineño’ and ‘Cimarron’ and Agave cupreata Trelease et Berger, 1915 by physical mapping of 5S and 18S rDNA from Agave tequilana ‘Azul’.
Plants were collected in the Denomination of Origin Zone for Agave tequilana ‘Azul’ and in southern Jalisco, México (municipality of Tolimán) for Agave angustifolia ‘Lineño’ and ‘Cimarron’ and in Miraval, Guerrero for Agave cupreata. Three accessions of each species and varieties were used in this work; the accessions were planted in pots containing a mixture of organic soil:sand:vermiculite (3:3:1) and kept under standard greenhouse conditions.
Elongating secondary root tips were treated with 2 mM 8-hydroxyquinoleine for 6 hours at 18 °C, in darkness. Later, root tips were fixed in ethanol:acetic acid (3:1) for 24 hours. Root tips were hydrolyzed with 1 N HCl for 15 minutes at 60 °C, transferred to Schiff’s reagent for 1 hour, and then to 1.8% propionic orcein to stain chromosomes (
A negative film was used to draw and measure the chromosome arms and the total genome length. The centromere position was obtained following
Root tips of each three accessions of Agave tequilana ‘Azul’, Agave angustifolia ‘Lineño’ and Agave angustifolia ‘Cimarron’ and Agave cupreata were collected early in the morning, pretreated with satured α-bromonaphthalene solution and kept in ice water overnight, then fixed in ethanol:acetic acid (3:1), for at least 12 hours and stored at -20 °C until use. Root tips were incubated in a pectolytic-enzyme mixture, containing 0.2% (w/v) pectolyase (Sigma, USA), 0.2% (w/v) cellulase Onozuka RS (Yakult, Japan), and 0.2% (w/v) cytohelicase (Sigma) in 10 mM citrate buffer (pH 4.5), at 37 °C for approximately 2 hours. Squash preparations were made in a drop of 45% acetic acid and frozen in liquid nitrogen; the cover slips were removed with a razor blade and slides were dehydrated in absolute ethanol and then air-dried. The best slides were stored at 2–3 °C for up to 1 month.
Total genomic DNA from Agave tequilana ‘Azul’ was extracted from fresh young leaves using the CTAB method (
5S and 18S rDNA probes were isolated with the High Pure Plasmid Isolation kit (Roche Diagnostics GmbH, Germany) and labeled with biotin-16-dUTP by nick translation according to the manufacturer’s instructions (Roche Diagnostics GmbH).
Slide pretreatment. Slides were incubated in RNase A (100 μg ml-1 in 2× SSC) for 1 hour at 37 °C, and washed with 2× SSC for 15 minutes. Then, the slides were incubated in 0.01 M HCl for two minutes and followed by treatment in pepsin (5 μg ml-1) in 0.01M HCl for 10 minutes at 37 °C. Afterwards, the slides were washed in 2× SSC for 10 minutes and incubated in 4% paraformaldehyde for 10 minutes at room temperature. Finally, the slides were dehydrated in ethanol series (70%, 90%, and absolute ethanol for 3 minutes each), and air-dried.
Probe hybridization. Hybridization was carried by using a mixture consisting of 20× SSC, formamide, 50% sodium dextran sulphate, 10% sodium dodecyl sulphate, and 25-50 ng/slide of each probe. DNA probes were denatured by heating the hybridization mixture at 70 °C for 10 minutes and then placing it on ice for at least 10 minutes. For each slide, 40 μl of the hybridization mixture were used. Slides were denatured at 80 °C for 5 minutes. The slides were then placed in a pre-warmed humid chamber and incubated overnight at 37 °C. Slides were washed at 37 °C in 2× SSC for 15 minutes, 0.1× SSC at 42 °C for 30 minutes, and 2× SSC at room temperature for 10 minutes.
Signal detection. Biotin-labeled probes were detected with streptavidin-Alexa Fluor546 conjugate (Life Technologies Corporation) and amplified with biotinylated goat-antistreptavidin (Vector Laboratories, USA). Chromosomes were counterstained with DAPI solution (1 μg ml-1), and one drop of Vectashield antifade (Vector Laboratories) was added before examination under a Leica DMRA2 microscope (Leica Microsystems, Germany) equipped with epifluorescent illumination and coupled to an Evolution QEi Camera (Media-Cybernetics, USA), and the images were analyzed with the Image-Pro software (Media-Cybernetics) and enhanced with Photoshop (Adobe Systems Incorporated, USA).
The partial amplification of 18S rDNA generated one band, which was cloned into electrocompetent Escherichia coli DH5α cells and a single clone was isolated, which after sequencing showed a fragment of 1424 bp (GenBank: KF159807) and a maximal identity of 100 % with Agave tequilana cultivar Azul (GenBank: GU980213.1) and Agave ghiesbreghtii K.Koch, 1862 voucher Chase 3467(K) (GenBank: HM640709.1) according to BLASTn analysis (nucleotide blast) at the NCBI database. The partial amplification of 5S rDNA generated one band, which was cloned into electrocompetent Escherichia coli DH5α cells and one clone was isolated, which after sequencing showed a fragment of 436 bp (GenBank: KF159808) and a maximal identity of 97% with Arabidopsis thaliana (Linnaeus, 1753) clone CIC YAC 9A12 and 9A5 5S ribosomal RNA gene (GenBank: AF198223.1), according to BLASTn analysis (nucleotide blast) at the NCBI database.
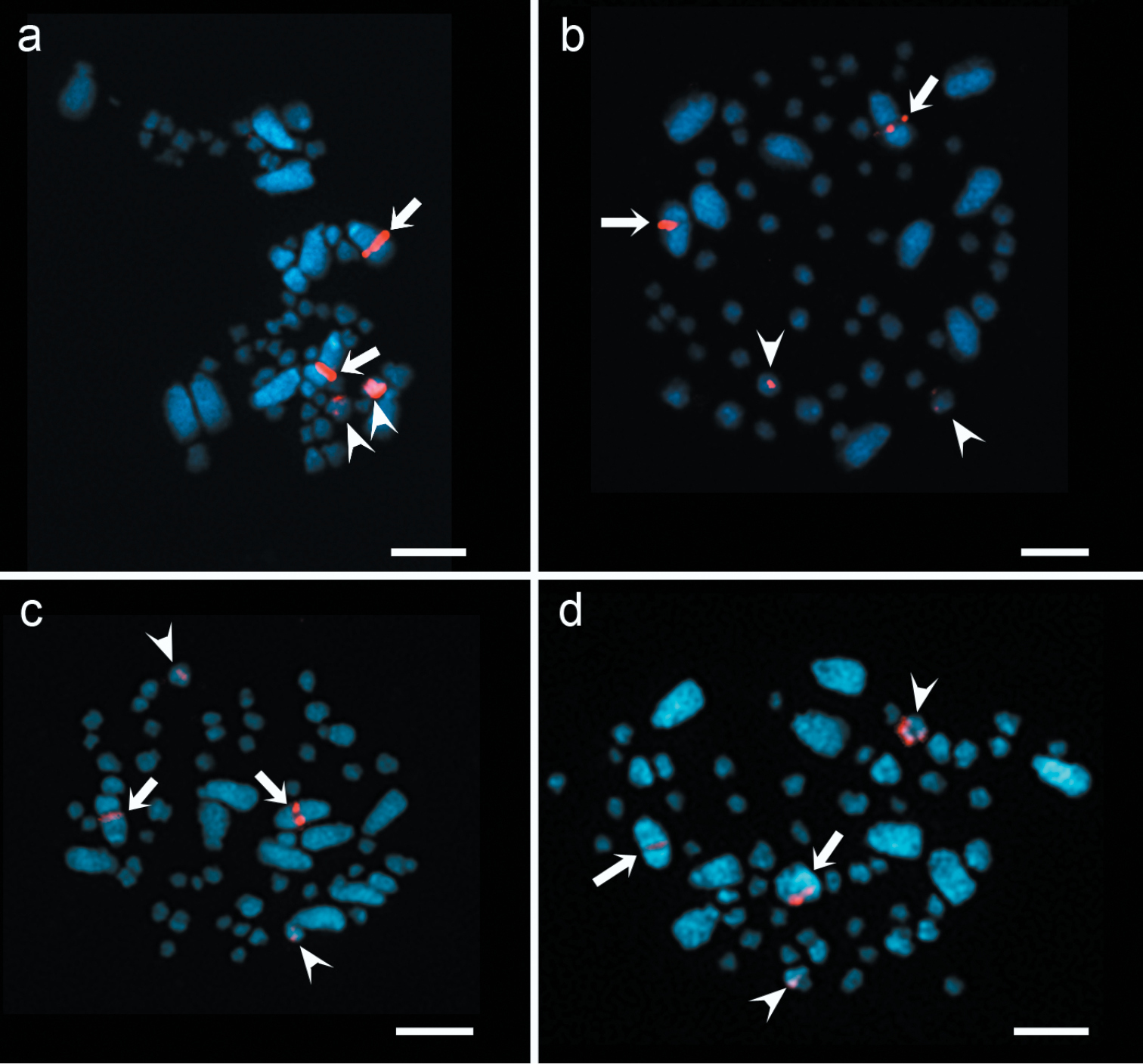
The physical mapping of 5S and 18S rDNA from Agave tequilana ‘Azul’ were investigated by fluorescent in situ hybridization (FISH) (Fig. 1). FISH experiments with both probes labeled with biotin and detected as a red signals, showed that the number of sites of rDNA were constant among all the species under study. 5S rDNA loci were located in both arms of small chromosome pair in each species (Fig. 1). The hybridization sites of cloned 18S rDNA were associated with the secondary constriction of a large chromosome pair in each species, being a subtelocentric chromosome pair in Agave tequilana ‘Azul’ and a telocentric chromosome pair in Agave cupreata and Agave angustifolia ‘Lineño’ and ‘Cimarron’ (Fig. 1).
FISH of 5S and 18S rDNA in Agave species. Two hybridization sites of 18S rDNA (arrows) and 5S rDNA (arrowheads) in: a Agave tequilana ‘Azul’ b Agave cupreata c Agave angustifolia ‘Lineño’ d Agave angustifolia ‘Cimarron’. Bars = 10 µm.
All the studied species were diploids with 2n = 2x = 60, confirmed by chromosome counting, considering the basic chromosome number x = 30 for the genus, and showed a bimodal karyotype with five pairs of large chromosomes and 25 pairs of small chromosomes. Karyotype analysis of Agave species is summarized in Table 1, and where it can be seen that all species showed different karyotypic formulae as well as a secondary constriction in one large chromosome pair; in Agave tequilana ‘Azul’ it was observed in pair 1, in Agave cupreata in pair 3, in Agave angustifolia ‘Lineño’ in pair 5 and in Agave angustifolia ‘Cimarron’ in pair 2.
Karyotypes in Agave species (2n = 2x = 60).
| Taxa and origin | Collector and voucher information | Karyotype formula | Secondary constriction |
|---|---|---|---|
| Agave angustifolia 'Cimarron' Tolimán, Jalisco State, México. 19°32'06"N, 103°53'44"W (DMS). | Rodríguez JM C |
42m + 4sm + 6st + 8t |
2t |
| Agave angustifolia 'Lineño' Tolimán, Jalisco State, México. 19°32'06"N, 103°53'44"W (DMS). | Rodríguez JM L |
48m + 2sm+ 2st + 8t |
2t |
| Agave cupreata Miraval, Guerrero State, México. 17°43'00"N, 99°45'00"W (DMS). | Trinidad RA 573 |
42m + 2sm + 8st + 8t |
2t |
| Agave tequilana 'Azul' CIATEJ, Jalisco State, México. 20°41'39"N, 103°20'47"W (DMS). | Rodríguez JM A, C, D |
42m + 12st + 6t |
2st |
† = Palomino et al. unpublished data.
‡ = Karyotype published by
§ = Karyotype published by
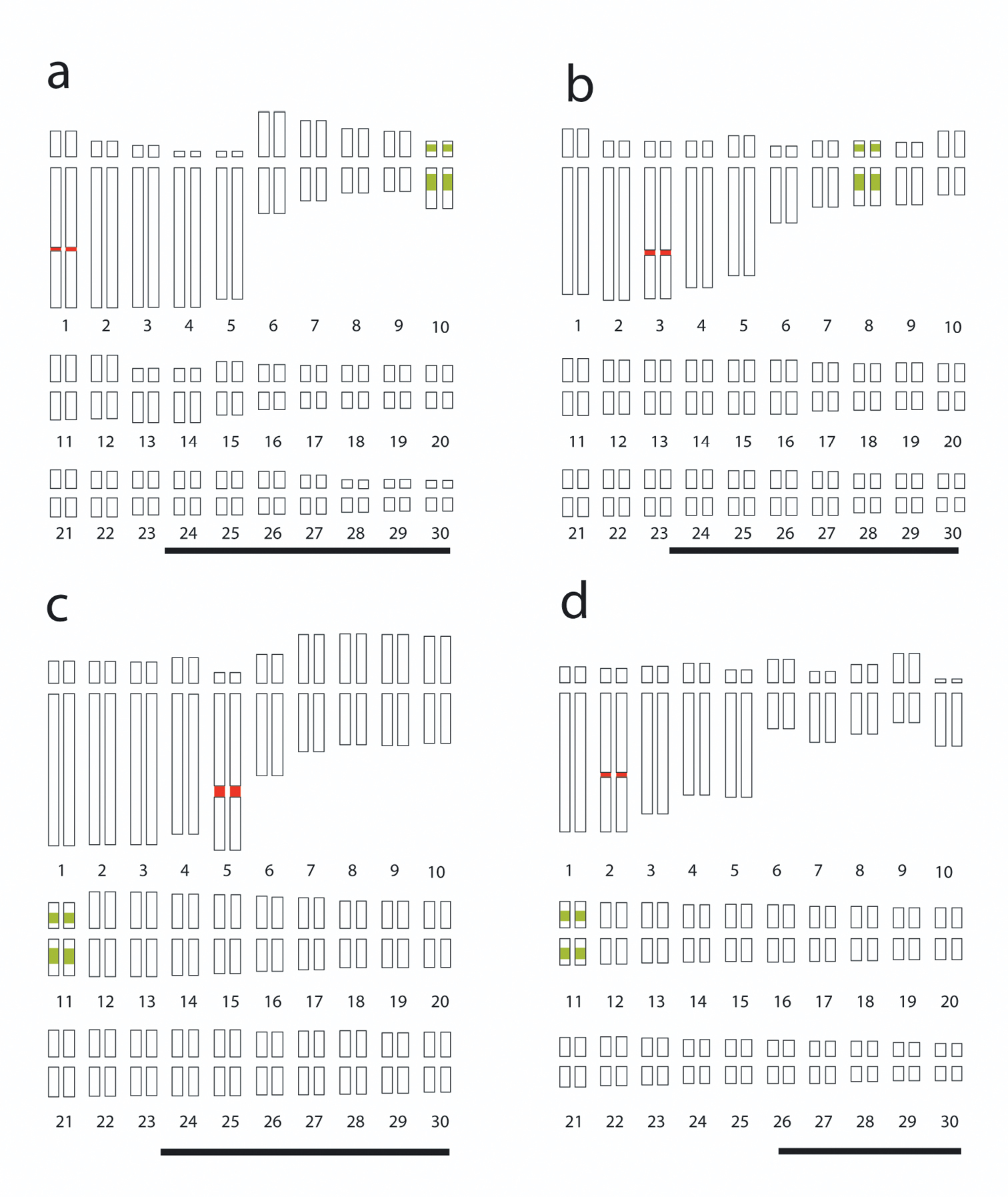
FISH data were integrated in idiograms, indicating the number and position of rDNA loci (Fig. 2). 5S rDNA loci always were located in a proximal region on both arms of a small chromosome in each species, whereas 18S rDNA loci always were located in the interstitial region of a large chromosome. Fig. 2a shows a hybridization signal of 18S rDNA in Agave tequilana ‘Azul’ on pair 1, while the 5S rDNA signals are on both arms of pair 10; in Agave cupreata (Fig. 2b), the hybridization signal of 18S rDNA is on pair 3, while the 5S rDNA signals are on both arms of pair 8; in Agave angustifolia ‘Lineño’ and ‘Cimarron’ (Fig. 2c-d), the hybridization signal of 18S rDNA is on pair 5 and 2, respectively, while the 5S rDNA signals are on both arms of pair 11 in both varieties.
Idiograms of Agave karyotypes showing the 5S (green) and 18S (red) rDNA loci. a Agave tequilana ‘Azul’ b Agave cupreata; c Agave angustifolia ‘Lineño’ d Agave angustifolia ‘Cimarron’. Bars = 10 µm.
Cytogenetic analysis showed the diploid chromosome number 2n = 60 in all species, which is in agreement with previous reports in the genus (
FISH with rDNA probes showed that loci of 18S and 5S rDNA in Agave species were located in different chromosomes and on similar position in all species; this finding suggests that the chromosomes bearing the rDNA loci are homeologous and the difference in numerical assignment is due to chromosomal rearrangements as mentioned before. 18S rDNA locus always was located in the interstitial region on the large arm of a large chromosome and associated to the secondary constriction, whereas the 5S rDNA loci were located in a proximal region on both arms of a small chromosome in all species. These results differ from
To the best of our knowledge, here we reported the number and location of rDNA loci in two species with no previous report, Agave cupreata and Agave angustifolia ‘Lineño’ and ‘Cimarron’ as well as a different locus of 5S rDNA in all species studied. Data of FISH analysis provides new information about physical mapping of rDNA in Agave and such identified sites can be useful as chromosome markers for chromosome identification in hybrids in breeding programs as well as in evolutionary studies.
Despite the great diversity of the genus Agave which includes 166 species, the physical mapping of rDNA or other molecular markers are scarce, since just about five species have been described. The different karyotype formulae found in all species indicated the presence of cytotypes and data of FISH of rDNA allowed the physical mapping of Agave cupreata and two new varieties of Agave angustifolia. This work provides new information about the position and number of rDNA loci in Agave species through comparative karyotype analysis, however, further cytogenetic research must be conducted to understand the evolution of this genus and develop breeding programs to preserve its biodiversity.
The authors would like to thank SEP-CONACYT-Mexico Project 24554, CONACYT, FOMIX-JAL Project 99210, who supported this research, and to Jose Manuel Rodriguez Dominguez for his technical assistance in field work. Also the authors would like to thank Dr. Ignacio del Real Laborde (Tequila Sauza, S. de R.L. de C.V.) for providing Agave tequilana plant material. VMGR is a graduate student and financially supported by CONACYT-Mexico (Reg. 45382).