(C) 2013 Patrícia Corrêa da Silva. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: da Silva PC, Nagamachi CY, dos Santos Silva D, Milhomem SSR, Cardoso AL, de Oliveira JA, Pieczarka JC (2013) Karyotypic similarities between two species of Rhamphichthys (Rhamphichthyidae, Gymnotiformes) from the Amazon basin. Comparative Cytogenetics 7(4): 279–291. doi: 10.3897/CompCytogen.v7i4.4366
The family Rhamphichthyidae includes three genera: Rhamphichthys Müller et Troschel, 1846, Gymnorhamphichthys M. M. Ellis, 1912 and Iracema Triques, 1996. From this family, only the species Rhamphichthys hanni Meinken, 1937 has had its karyotype described. Here, we describe the karyotypes of two additional Rhamphichthys species: Rhamphichthys marmoratus Castelnau, 1855 from the Reserva de Desenvolvimento Sustentável Mamirauá, Amazonas state and Rhamphichthys prope rostratus Linnaeus, 1766 from Pará state, both in Brazil. Our karyotypic analyses demonstrated that the diploid number is conserved for the genus (2n = 50), but the karyotypic formulas (KFs) differed between Rhamphichthys marmoratus (44m/sm+6a) and Rhamphichthys prope rostratus (42m/sm+8a). In both species, the constitutive heterochromatin (CH) was located in the centromeric region of most chromosomes. Large heterochromatic blocks were found on the long arms of pairs 4 and 14 in Rhamphichthys marmoratus and on chromosomes 3, 4 and 19 in Rhamphichthys prope rostratus, which also has a heteromorphism in chromosome pair 1. The CH was DAPI positive, indicating that it is rich in AT base pairs. The Nucleolus Organizer Region (NOR) showed staining at a single location in both species: the long arm of pair 1 in Rhamphichthys marmoratus and the long arm of pair 12 in Rhamphichthys prope rostratus, where it showed a size heteromorphism. CMA3 staining coincided with that of Ag-NOR, indicating that the ribosomal genes contain interspaced GC-rich sequences. FISH with an 18S rDNA probe confirmed that there is only one NOR site in each species. These results can be used as potential cytogenetic markers for fish populations, and comparative analysis of the karyotypes of Hypopygus Hoedman, 1962, Rhamphichthys and Steatogenys Boulenger, 1898 suggests that the first two genera diverged later that the third.
Gymnotiformes, Rhamphichthyidae, Cytogenetics, FISH
The family Rhamphichthyidae comprises three genera: Rhamphichthys Müller et Troschel, 1846, with eight described species, Gymnorhamphichthys Ellis, 1912, with six species, and Iracema Triques, 1996, with only one species (
Species of Rhamphichthyidae (According to
| Species | Locality |
|---|---|
| Gymnorhamphichthys hypostomus Ellis, 1912 | São Joaquim, Bolivia |
| Gymnorhamphichthys rondoni Miranda Ribeiro, 1920 | 17 de Fevereiro River, Amazonas, Brazil |
| Gymnorhamphichthys petiti Géry et Vu-Tân-Tuê, 1964 | Bananal Island, Araguaia River, Brazil |
| Gymnorhamphichthys rosamariae Schwassmann, 1989 | Negro River, Amazonas, Brazil |
| Gymnorhamphichthys bogardusi Lundberg, 2005 | Orinoco River, Delta Amacuro State |
| Gymnorhamphichthys britskii Carvalho et al., 2011 | Paraná- Paraguay System |
| Iracema caiana Triques, 1996 | Jauaperi Beach, Negro River, Amazonas, Brazil |
| Rhamphichthys apurensis Fernández-Yépez, 1968 | Bucaral River, a tributary of Apure River, Venezuela |
| Rhamphichthys atlanticus Triques, 1999 | Viana Lake, Amazonas, Brazil |
| Rhamphichthys drepanium Triques, 1999 | Janauari Lake, confluence of the Negro and Solimões Rivers, Amazonas, Brazil |
| Rhamphichthys hahni Meinken, 1937 | Paraná River basin, next to Corrientes, Argentina |
| Rhamphichthys lineatus Castelnau, 1855 | Ucayali River basin, Peru |
| Rhamphichthys longior Triques, 1999 | Paru Lake, confluence of the Trombetas River, Para, Brazil |
| Rhamphichthys marmoratus Castelnau, 1855 | Araguaia River, Brazil; Ucayali River, Peru |
| Rhamphichthys rostratus Linnaeus, 1766 | South America |
The species of Rhamphichthys have a long and narrow body, a long tubular snout, no teeth in the jaw, and an anal fin with more than 300 rays. They are slow swimmers and spend most of their time at the bottoms of rivers (
The phylogeny of the Gymnotiformes proposed by
Relatively few cytogenetic studies have been performed in Gymnotiformes. According to Oliveira et al.(2009), only 48 species of this order have had their karyotypes described. The genera Gymnotus Linnaeus, 1758 and Eigenmannia Jordan et Evermann, 1896 have the most available information on their karyotypic diversity (
In Rhamphichthyoidea, the available chromosome information comes from only six species (Table 2): Hypopomus artedi Kaup, 1856 with diploid number (2n) = 38, Fundamental Number (FN) = 70 and Karyotypic Formula (KF) = 32m/sm+6st/a; Hypopygus lepturus Hoedman, 1962with 2n = 50, FN = 86 and KF = 36m/sm+10st+4a; Brachyhypopomus brevirostris Steindachner, 1868, with 2n = 36, FN = 42 and KF = 6m/sm+30st/a (
A review of the cytogenetic information in Rhamphichthyoidea from
| Family / Species | 2n | KF | Sex system | CB | NOR | References |
|---|---|---|---|---|---|---|
| Hipopomidae | ||||||
| Hypopomus artedi Kaup, 1856 | 38 | 32m-sm / 6st-a | Absent | - | - |
|
| Brachyhypopomus brevirostris Steindachner, 1868 | 36 | 6m-sm / 30st-a | Absent | - | - |
|
| Brachyhypopomus pinnicaudatus (Hopkins, 1991) | 41♂ / 42♀ | 1m/41a♂ / 42a♀ | X1X2Y | Centromeric region of most chromosomes | Multiple |
|
| Hypopygus lepturus Hoedeman, 1962 | 50 | 36m-sm / 14st-a | Absent | - | - |
|
| Steatogenys elegans (Steindachner, 1880) | 50 | 12m-sm/ 38st-a | ZZ/ZW | Centromeric region of all chromosomes and interstitial (1q and 2 blocks in Wq) | Single |
|
| Steatogenys duidae (La Monte, 1929) | 50 | 50 m-sm | Absent | Centromeric and pericentromeric region of all chromosomes and interstitial (2q , 3q, 5q and 7q) | Single |
|
| Rhamphichthyidae | ||||||
| Rhamphichthys hahni (Meinken, 1937) | 50 | 44m-sm / 6a | Absent | Centromeric region of most chromosomes and blocks of CH in three chromosomes (SM) | Single |
|
| Rhamphichthys marmoratus Castelnau, 1855 | 50 | 44m-sm / 6st-a | Absent | Centromeric region of most chromosomes and interstitial blocks (4q and 14p) | Single | Present work |
| Rhamphichthys prope rostratus (Linnaeus, 1766) | 50 | 42m-sm / 8a | Absent | Centromeric region of most chromosomes and interstitial blocks (3q, 4q and 19p) | Single | Present work |
In the present work, we studied the karyotypes of two species of Rhamphichthys from the Amazon region in an effort to better define the boundaries between the species, and compared our findings with those from the single previously described species of Rhamphichthys to better understand the phylogenetic relationships in this genus.
Fishes were collected using a bioamplification device that detects electric fields and translate them into sounds (
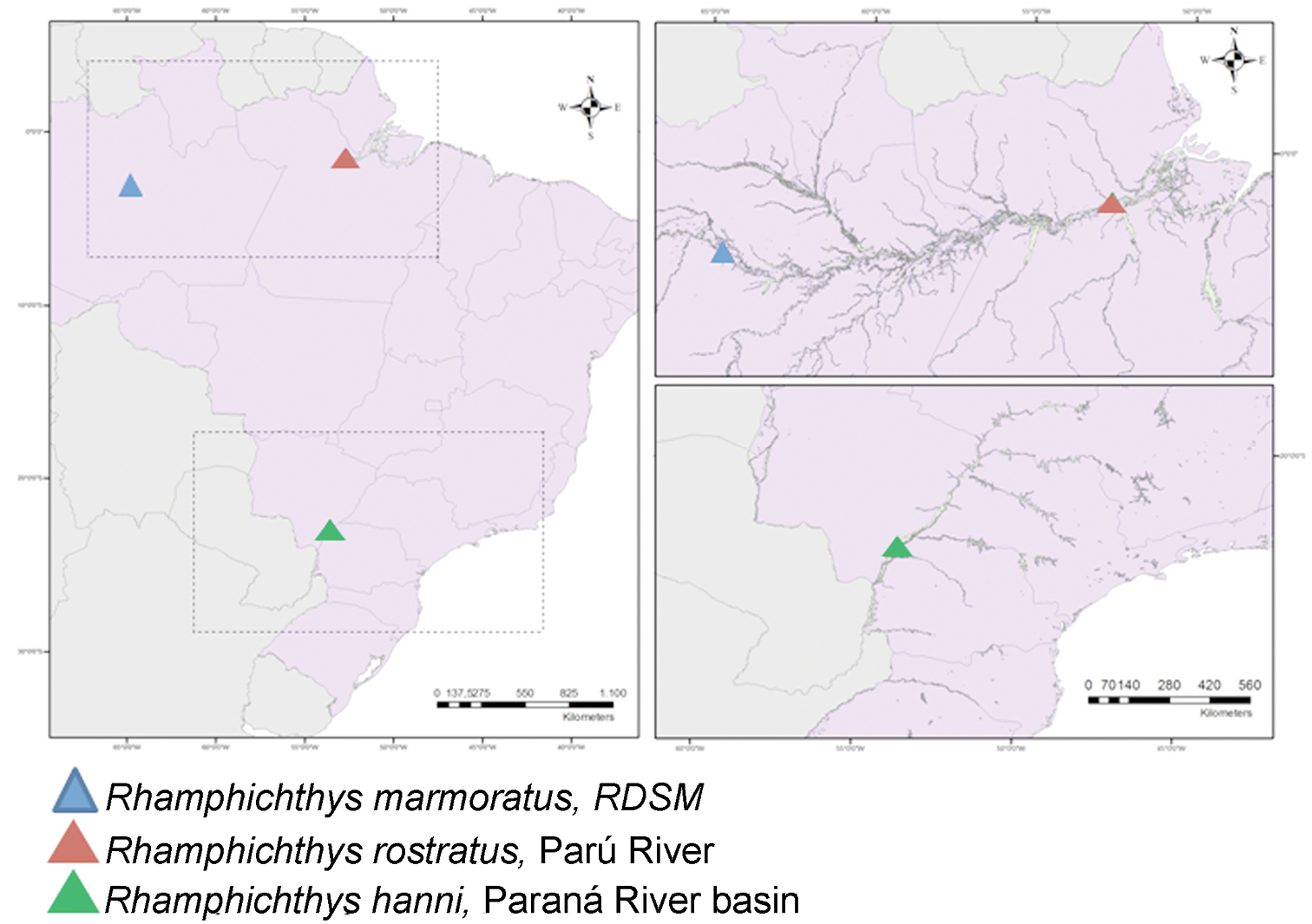
A map with the location of the Rhamphichthys species with cytogenetic descriptions. Rhamphichthys marmoratus and Rhamphichthys rostratus were analyzed in the present work.
Metaphase chromosomes were obtained according to the method described by
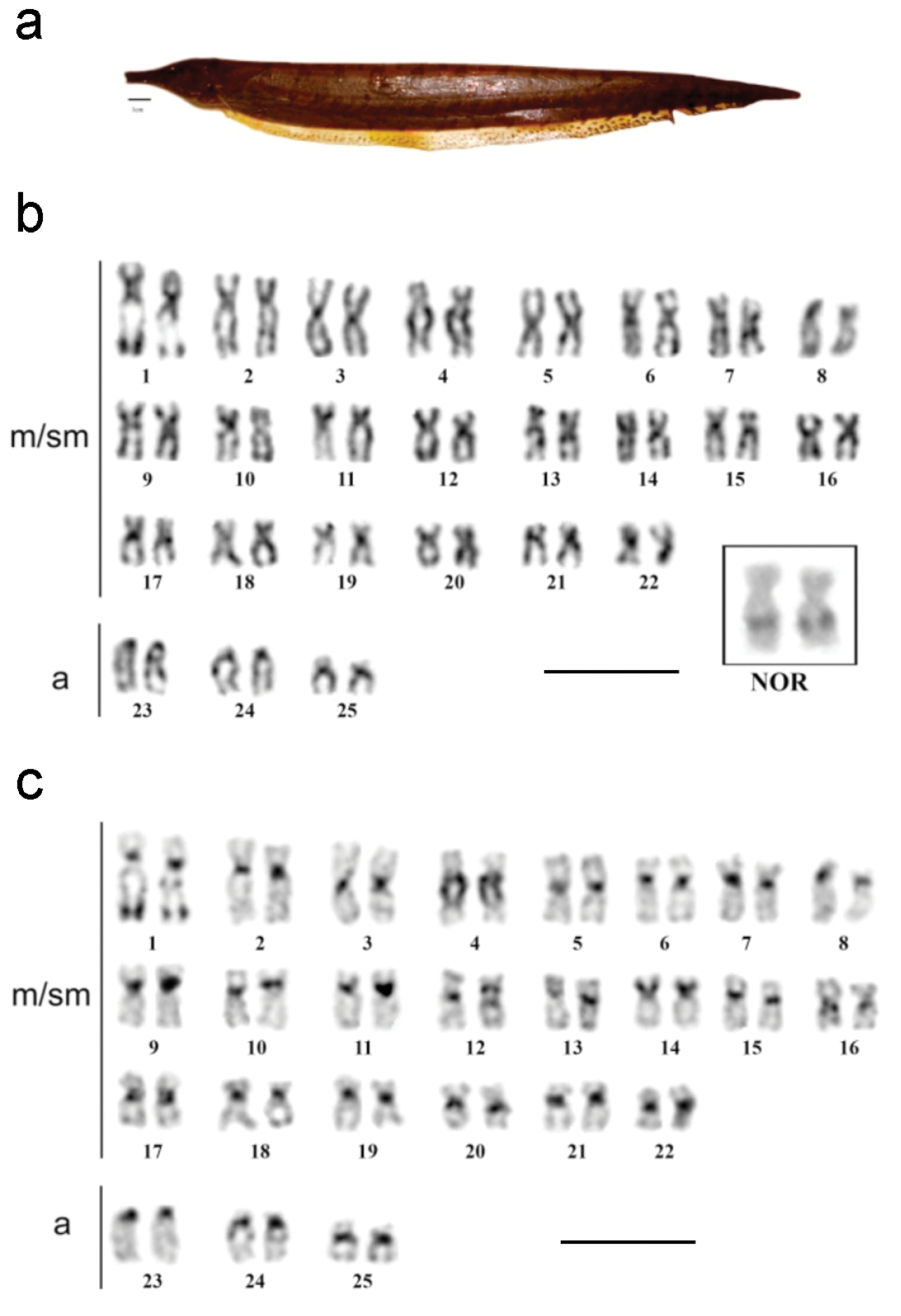
All samples of Rhamphichthys marmoratus (Fig. 2) had 2n = 50 and a karyotypic formula (KF) consisting of 44 metacentric/submetacentric (m/sm) and 6 acrocentric chromosomes (Fig. 2a), with no evidence of any sex-determination chromosome system. Ag-NOR staining showed that the NOR is located in the interstitial region of the long arm of pair 1, in a secondary constriction (Fig. 2b, box). Constitutive heterochromatin (CH) was found in the centromeric regions of all chromosomes (Fig. 2c). Pair 4 was notable for a large heterochromatic block running from the proximal region across most of the long arm, while pair 14 had a CH block covering most of its short arm. CH was also found in the distal region of the long arm of pair 1 (Fig. 2c). DAPI fluorochrome banding coincided with positive C-banding in all centromeres, and was especially strong in pairs 4 (Fig. 3a). The CMA3 fluorochrome banding localized to the same region as the NOR, suggesting that this region is GC-rich (Fig. 3b). FISH with 18S rDNA probes confirmed that the NOR is located in the interstitial region of the long arm of pair 1 (Fig. 3c).
a Rhamphichthys marmoratus b Giemsa stained karyotype with the NOR bearer pair into the box c C-banded sequenced karyotype (m/ms- metacentric/submetacentric, a- acrocentric). Scale bar: a) 1 cm, b) and c) 10 μm.
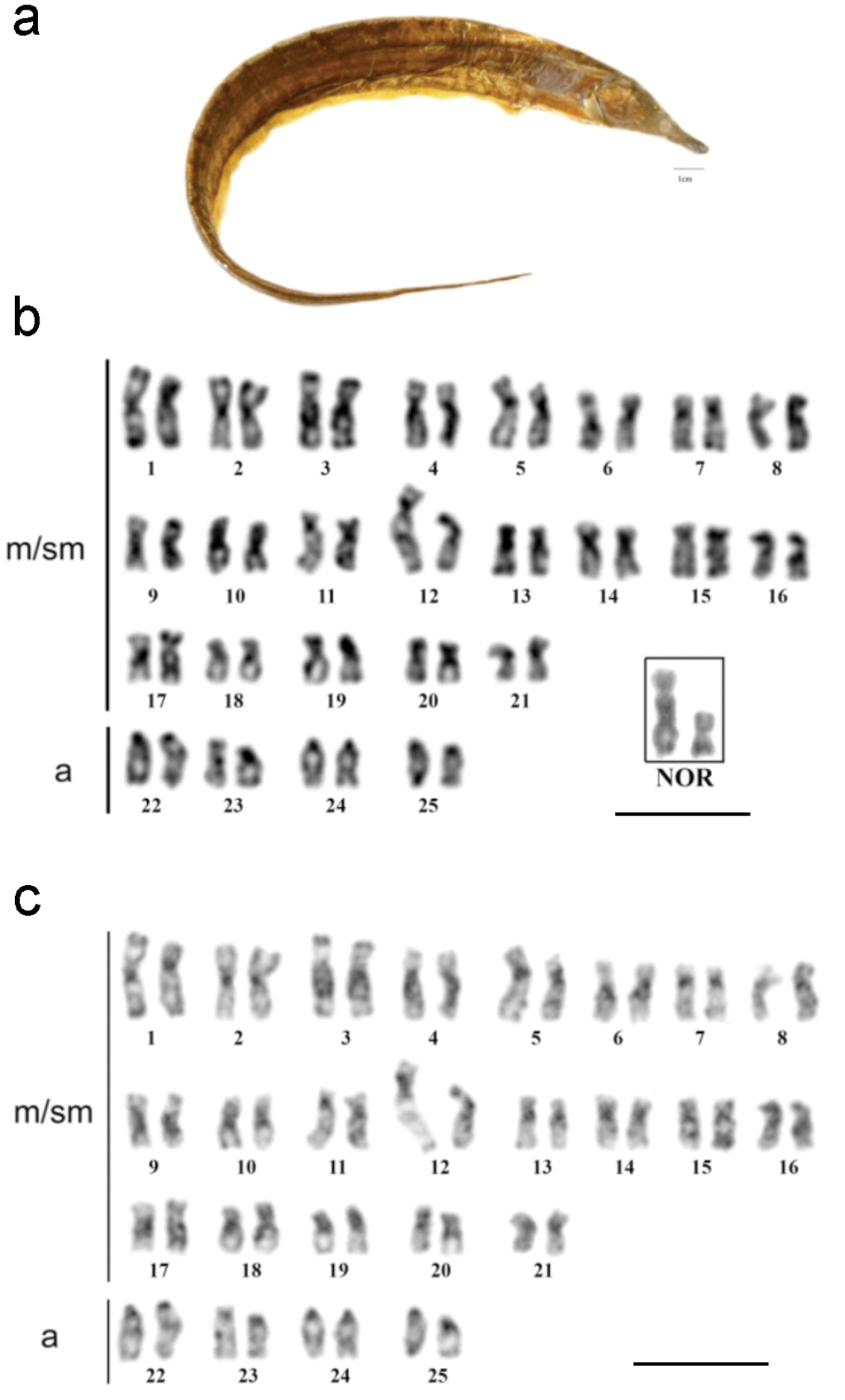
a Rhamphichthys prope rostratus b Giemsa stained karyotype with the NOR bearer pair into the box c C-banded sequenced karyotype; (m/ms- metacentric/submetacentric, a- acrocentric). Scale bar: a) 1 cm, b) and c) 10 μm.
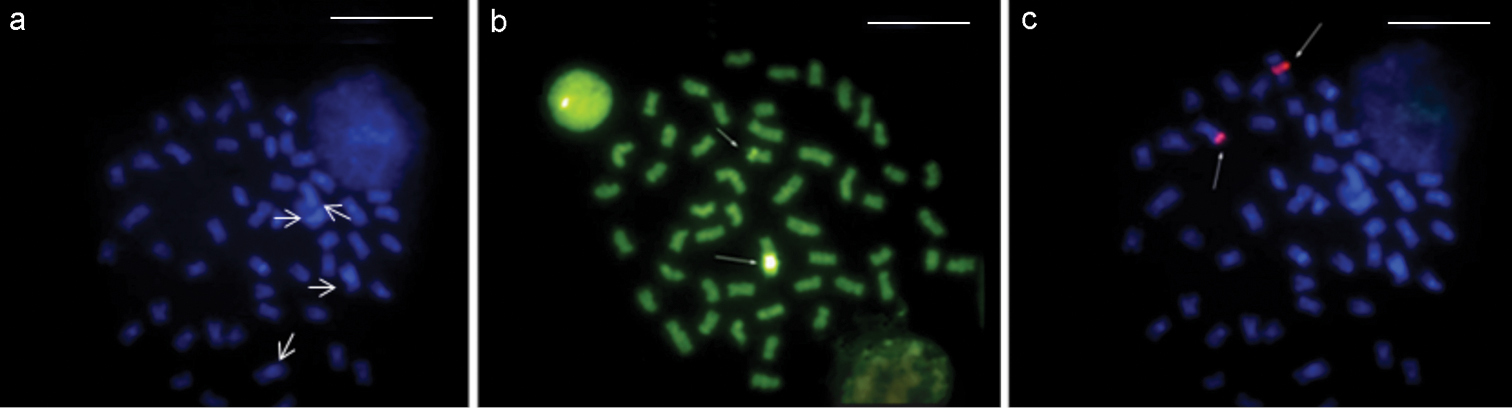
Rhamphichthys prope rostratus (Fig. 4a) had 2n = 50 and a KF of 42m/sm+8a, with no evidence of a sex-determination system (Fig. 4b). Ag-NOR staining was noted in the interstitial region of the long arm of pair 12 (Fig. 4b, box). CH was found in the pericentromeric regions of most chromosomes, and large CH blocks were found in the proximal regions of the long arm of pairs 3, 4 and 9. Pair 1 had a heteromorphism in both males and females, probably because of a heterochromatin block, as did pair 12 (Fig. 4c). DAPI banding was positive in the CH regions, suggesting that these regions are AT-rich (Fig. 5a). CMA3 banding showed size differences between the homologs, suggesting the presence of a size difference in this GC-rich region (Fig. 5b). Finally, FISH against the 18S rDNA hybridized to the same region that was positive for Ag-NOR staining (Fig. 5c).
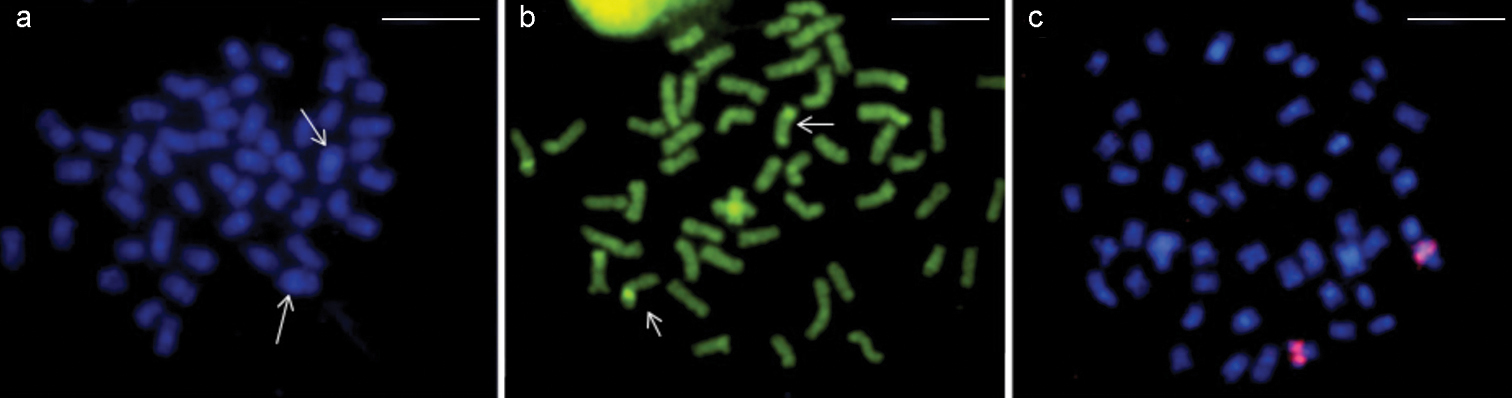
Rhamphichthys marmoratus - a DAPI staining. Arrows: pair 4 with a large CH block b CMA3 staining, arrows designate NOR pair c FISH with rDNA probe. Scale bar: 10 μm.
Rhamphichthys rostratus - a DAPI staining, arrows designate pairs 3 and 4 with large CH blocks b CMA3 staining, arrows designate NOR pair c FISH with rDNA probe. Scale bar: 10 μm.
Both Rhamphichthys marmoratus and Rhamphichthys prope rostratus had 2n = 50, but differed in their KFs, with Rhamphichthys marmoratus having 44m/sm+6a and Rhamphichthys prope rostratus having 42m/sm+8a. Previously, Rhamphichthys hanni was described as having 2n = 50, but 20m+24sm+6a (
The CH in Rhamphichthys prope rostratus and Rhamphichthys marmoratus is AT-rich (i.e., DAPI banding-positive), which is consistent with other species of Gymnotiformes (
The NOR was found on a secondary constriction and stained positive with CMA3 as previously observed on other species (Pendáset al. 1993, Fernandes et al. 2005,
The phylogeny proposed by
However
The available cytogenetic information on Gymnotiformes may be sparse (of eight species of this genus, only three have had their karyotypes analyzed), but the existing data show an important variability in this group. More cytogenetic investigations on the family Rhamphichthyidae are warranted, as they will help us better understand the chromosomal evolution of these fishes for use in other fields of science, and assist us in defining the boundaries of the Rhamphichthys species.
The authors thank the following: the IDSM (Instituto de Desenvolvimento Sustentável Mamirauá) for allowing the sample collectors into the reservoir and providing logistical support; FAPESPA, CNPq and CAPES for financial support; the fishes staff members in the Cytogenetics Laboratory of Universidade Federal do Para for supporting the collection of field samples and critically reading this paper; and MSc. Anderson Gomes for help with drawing the map and providing other contributions.