Citation: Steinberg ER, Nieves M, Mudry MD (2014) Multiple sex chromosome systems in howler monkeys (Platyrrhini, Alouatta). Comparative Cytogenetics 8(1): 43–69. doi: 10.3897/CompCytogen.v8i1.6716
In light of the multiple sex chromosome systems observed in howler monkeys (Alouatta Lacépède, 1799) a combined cladistic analysis using chromosomal and molecular characters was applied to discuss the possible origin of these systems. Mesoamerican and South American howlers were karyologically compared. FISH analysis using the chromosome painting probes for the #3 and #15 human chromosomes was applied to corroborate the homeology of the sexual systems. We found that the HSA3/15 syntenic association, present in the sex chromosome systems of South American Howlers, is not present in those of Mesoamerican ones. The autosomes involved in the translocation that formed the sexual systems in the Mesoamerican and South American species are different, thus suggesting an independent origin. Parsimony analysis resolved the phylogenetic relationships among howler species, demonstrating utility of the combined approach. A hypothesis for the origin of the multiple sex chromosome systems for the genus is proposed.
Multiple sex chromosome systems, combined phylogenetic analysis, FISH, Neotropical Primates, cytochrome b
Howler monkeys (genus Alouatta Lacépède, 1799 of the family Atelidae) exhibit one of the widest geographic distributions recorded to date for Neotropical Primates. Their distribution extends from southern Mexico to northern Argentina (
In this context, and to contribute to the description of the phylogenetic relationships in the genus, several authors have proposed that chromosomal data can also be used as phylogenetic markers, since they are inherited as mendelian characters and are conserved within species (
In primates, different researchers in the last three decades have proposed chromosomal speciation as a probable evolutionary mechanism to explain the diversity observed in living species (
Cytogenetic characteristics of howler monkeys (Alouatta).
| Species | 2N | Sex Chromosome Systems | References |
|---|---|---|---|
| Alouatta belzebul | ♀50 ♂49 | X1X1X2X2/X1X2Y | |
| Alouatta seniculus seniculus | ♀♂47 to 49 |
XY | |
| Alouatta seniculus stramineus | ♀♂47 to 49 |
X1X1X2X2/X1X2Y1Y2 | |
| Alouatta seniculus arctoidea | ♀44 ♂45 |
X1X1X2X2/X1X2Y1Y2 | |
| Alouatta sara | ♀♂ 48 to 51 |
X1X1X2X2/X1X2Y | |
| ♀♂50 | X1X1X2X2/X1X2Y1Y2 | ||
| Alouatta macconnelli | ♀♂ 47 to 49 |
X1X1X2X2/X1X2Y1Y2 | |
| Alouatta caraya | ♀♂52 | XX/XY | |
| X1X1X2X2/X1X2Y1Y2 | |||
| Alouatta palliata | ♀♂56 | XX/XY | |
| ♀54 ♂53 | X1X1X2X2/X1X2Y | ||
| Alouatta pigra | ♀♂58 | X1X1X2X2/X1X2Y1Y2 | |
| Alouatta guariba guariba | ♀50 ♂49 | XX/XY | |
| ♂49 | X1X2Y | ||
| ♀50♂49 | X1X1X2X2X3X3/X1X2X3Y1Y2 | ||
| Alouatta guariba clamitans | ♀46 ♂45 | XX/XY | |
| X1X1X2X2/X1X2Y | |||
| X1X1X2X2X3X3/X1X2X3Y1Y2 | |||
| Alouatta nigerrima | ♀50 | XX | |
| Alouatta coibensis | ND | ND | --- |
† These differences are due to the presence of microchromosomes (1 to 3 per nuclei);
‡ Differences due to a variation in microchromosome number between sexes. ND: not yet cytogenetically characterized.
§ Meiotic studies performed to corroborate the sex chromosome system.
The phylogenies proposed so far for Alouatta have used either molecular markers (γ1-globin (
In the present contribution, howler species were karyologically compared and FISH analyses were carried out to corroborate the homeology of the sex chromosome systems among them. Using these data and molecular data obtained from the literature, a phylogenetic analysis combining them in a single matrix was performed.
Sampled specimens: A total of 29 adult specimens of both sexes of four species of howlers, both from captivity as well as from the wild within their natural geographical distribution, were analyzed: Alouatta caraya (9 ♂ and 6 ♀), Alouatta guariba clamitans (1 ♂), Alouatta pigra (6 ♂ and 5 ♀) and Alouatta palliata (2 ♂).
The origin of the animals was as follows:
Argentina
Alouatta caraya, 1 ♂ from Corrientes Zoo, Corrientes; 1 ♂ from Ecological Park “El Puma”, Misiones; 1 ♂ and 2♀ from Mendoza Zoo, Mendoza; 6 ♂ and 4 ♀ from the Black Howler Monkey Reeducational Center, La Cumbre, Córdoba.
Alouatta guariba clamitans, 1 ♂ from “Güira-Oga”, Misiones.
Mexico
Alouatta pigra: 4 ♂ and 4 ♀ were sampled in the wild in Campeche, Yucatán Península; 2 ♂ and 1 ♀ from San Juan de Aragón Zoo, Mexico City.
Alouatta palliata: 1 ♂ from San Juan de Aragón Zoo, Mexico City; 1 ♂ from Chapultepec Zoo, Mexico City.
Chromosome preparation: Peripheral blood samples were collected from all animals with previously heparinized disposable syringes. Lymphocytes were cultured for 72 h at 37 °C following
Analysis of homeologies: For Alouatta caraya and Alouatta guariba clamitans, the homeologies with human chromosomes and the homeologies with the other South American howlers are well known (
FISH analysis with human chromosome painting probes #3 and #15 was used as a tool to confirm the identity of the sex chromosome systems in howlers. Whole chromosome painting probes for human chromosomes #3 (red), #15 (green), #21 (green), X (green) and Y (red) (PCT3 Cy3, PCT15 FITC, PCT21 FITC, PCTX FITC, PCTY Cy3, LEXEL S.R.L., Buenos Aires, Argentina) were used for FISH analysis on the metaphases of Alouatta pigra, Alouatta caraya, Alouatta guariba clamitans and Alouatta palliata. Homo sapiens (HSA) metaphases were used as a positive control of hybridization. The HSA3/21 syntenic association, considered ancestral in mammals and conserved in most primate species (
FISH was performed according to the supplier’s instructions (LEXEL S.R.L., Buenos Aires, Argentina). Slides were counterstained with DAPI (Sigma) and analyzed with a Leica DMLB fluorescence microscope. Chromosome images were obtained with a Leica DFC 340 FX camera. Images were processed with Image Pro-Plus 4.5 (Media Cybernetics Inc.).
Our results were compared with those previously described (
Chromosomal dataset: We used data obtained from the comparisons of G-banding patterns and the analysis of chromosomal syntenic associations, both from the present study and from previous reports (
Human chromosome syntenic association considered as characters and used to construct the binary matrix of chromosomal homeologies among howler monkeys (modified from
| 1. 1p21-pter/1p12-21 2. 5q31.3-qter/ 7p22; q11 q21 3. 5pter-q31.2/5q31.3-qter 4. 2pter q12/16q 5. 4q31.3-qter/4q23-q31.2 6. 4q23-q31.2/4pter-q22 7. (10q/16p)3 8. 6 9. 8p/18 10. 15q21.3-q24/15q13-q21.2 11. 15q11 q13; q25 qter 12. 7p21 p11; q11 q21; q22 qter 13. 8q 14. 12 15. 11 16. 13 17. 9 18. 3pter p24; p21 p12; q12 q13; q27 qter 19. 3p24 q21; q13 q26 20. 1q32 qter 21. 1q21 q31 22. 3p12/21 23. 10p 24. 22 25. X 26. Y 27. 5pter-q31.2 28. (5q31.3-qter/7p22; q11 q21)2 29. 1p21-pter 30. 1p12-21 31. 4q31.3-qter 32. 4q23-q31.2/15q13-q212 33. 4pter-q22 34. 14/15q21.3-q24 35. (10q/16p)2/(10q/16p)1 |
36. 15q11 q13; q25 qter/Y 37. 2pter q12 38. 16q 39. 3p24 q21; q13 q26/ 15q11 q13; q25 qter 40. 11/5pter-q31.2 41. 5pter-q31.2/7p22; q11; q21 42. 12/9 43. 1p21-pter/2pter q12 44. 16q/4pter-q22 45. 22/14 46. 2q13 qter/20 47. 2q13 qter/4q23-q31.2 48. 8q/2q13 qter 49. 7p22; q11; q21/8q 50. 7p22; q11; q21/8q 51. 17/2pter q12/12 52. 2pter q12/12 53. 1q32 qter/11 54. 1q32 qter/(11/5pter-q31.2)2 55. (11/5pter-q31.2)2 56. 18/14 57. 3pter p24; p21 p12; q12 q13; q27 qter/15q21.3-q24 58. 3pter p24; p21 p12; q12 q13; q27 qter/15q21.3-q24/16q 59. 15q21.3-q24/16q 60. 17/10p 61. 17/10p/19 62. 10p/19 63. 22/20 64. 22/20/1q21 q31 65. 20/1q21 q31 66. (11/5pter-q31.2)3 67. 1p12-21/8p 68. 7p21 p11; q11 q21; q22 qter /14/15q21.3-q24 |
69. (10q/16p)2 70. 11/(10q/16p)2 71. 10q/16p 72. 10q/16p/4pter-q22 73. 10p/10q/16p/4pter-q22 74. 10p/10q/16p 75. 19/13 76. 19/22 77. 22/1p21-pter 78. 19/22/1p21-pter 79. 2q13 qter/4q23-q31.2 80. 6/1p12-21 81. Y/15q11 q13; q25 qter/ 3p24 q21; q13 q26 82. 1p12-21/5pter-q31.2/7p22; q11; q21/5q31.3-qter/ 7p22; q11 q21 83. 9/22 84. 17/11 85. 3pter p24; p21 p12; q12 q13; q27 qter/8p 86. 15q21.3-q24/1q32 qter 87. 16q/15q21.3-q24/1q32 qter 88. 4pter-q22/1p12-21 89. 14 90. 2pter q12/4pter-q22 91. 15q13-q21.2/7p22; q11; q21/5q31.3-qter/ 7p22; q11; q21 92. 6/15q21.3-q24 93. 14/1p12-21 94. 6/15q21.3-q24/14/1p12-21 95. 17/8p/18 96. 22/10q/16p 97. 2q13 qter/11 98. (10q/16p)2/1q21 q31/20 99. Y/7 |
/: separates the chromosomal segments that constitute an association. ()n: n = number of repeats in the segment.
Molecular dataset: The sequences available in GenBank for the same species used in the G-banding pattern and FISH comparisons were taken into to choose the molecular marker. The only molecular marker that fullfiled all the requirements was cyt b. The sequences used were (Genbank Accession Numbers): Alouatta belzebul (AY374348.2), Alouatta caraya (AY374359.2), Alouatta seniculus arctoidea (AY065886.1), Alouatta sara (AY065887.1), Alouatta macconnelli (AY065888.1), Alouatta guariba guariba (AY065899.1), Alouatta guariba clamitans (DQ679782.1), Alouatta pigra (AY065884.1), Alouatta palliata (AY065879.1) (
Phylogeny: A Maximum Parsimony phylogeny using the exhaustive search option was obtained with PAUP 4.0 software (Phylogenetic Analysis Using Maximum Parsimony, (
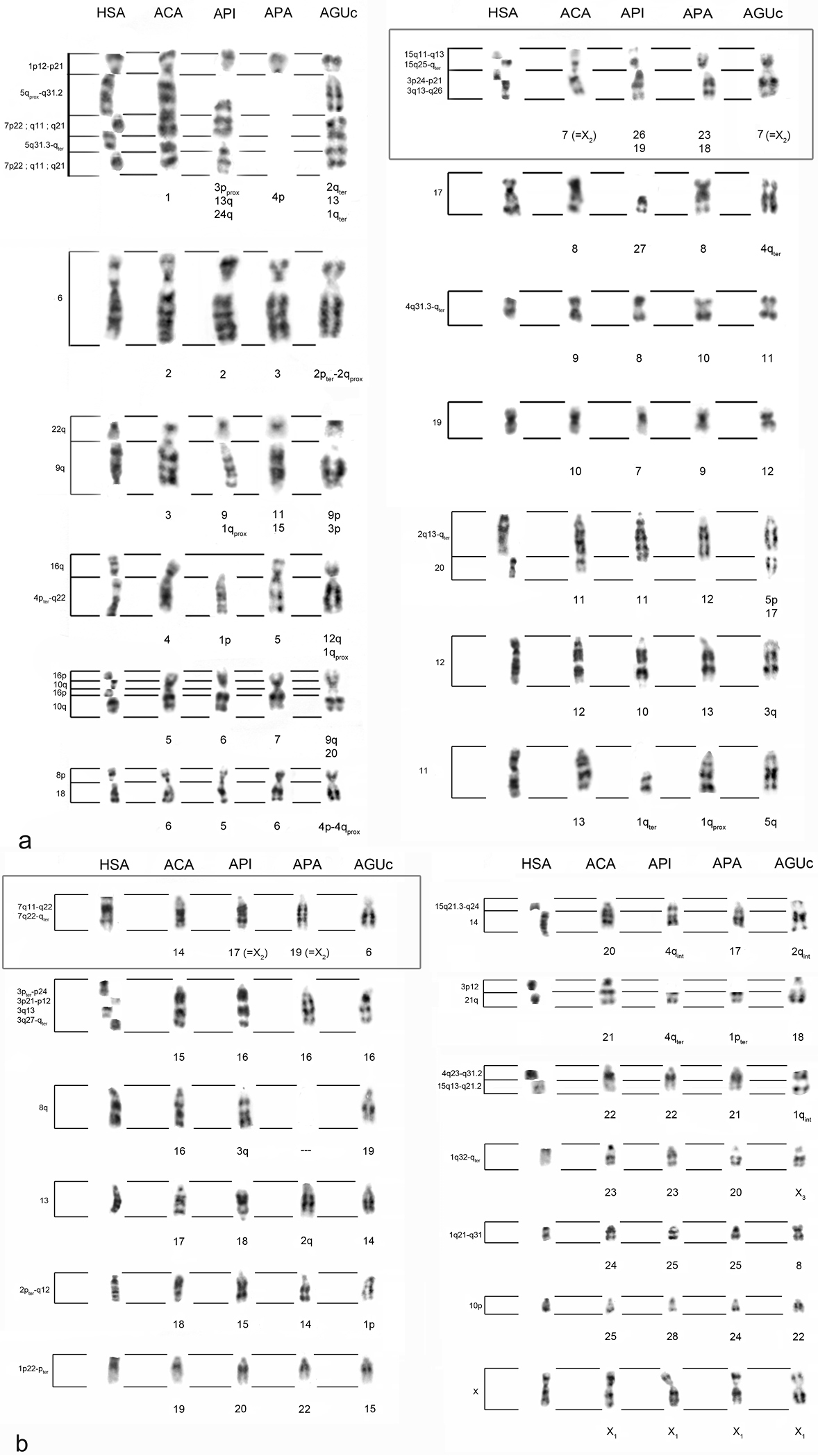
Karyological analysis: The cytogenetic characterization of the Alouatta specimens showed diploid numbers, sex chromosome systems and G-bandings patterns in agreement with the ones previously described for each species. Figures 1a and 1b show all the comparisons performed.
Comparison of Homo sapiens (HSA), Alouatta caraya (ACA), Alouatta pigra (API), Alouatta palliata (APA) and Alouatta guariba clamitans (AGUc) G-banded chromosomes, taking Alouatta caraya’s karyotype as reference. On the left, human chromosomal bands with homeology for its corresponding ACA chromosome segment are indicated. The boxes highlight the homeologies of the autosomes involved in the sex chromosome systems in these species a Comparison for ACA chromosomes #1 to #13 b Comparison for ACA chromosomes #14 to X1
Chromosomal homeologies between Alouatta caraya and Alouatta palliata: The chromosomal rearrangements that could explain the homeologies were grouped in two categories: 1) Alouatta palliata chromosomes with no rearrangements with respect to Alouatta caraya chromosomes: 3, 5, 6, 7, 8, 9, 10, 13, 14, 16, 17, 19, 20, 21, 22, 24, 25 and X1; 2) Alouatta palliata chromosomes with more than one rearrangement with respect to Alouatta caraya chromosomes: 1, 2, 4, 11, 12, 15, 18 and 23. No homeologies were allocated for Alouatta palliata chromosome 26 and chromosome arms 4q and 2p using the level of resolution of the classical cytogenetic techniques applied. The rearrangements detected between the Alouatta caraya and Alouatta palliata karyotypes included at least seven fissions/fusions, two paracentric inversions and one deletion. Alouatta caraya chromosome 7 (X2 in males) shares homeology with two Alouatta palliata chromosome pairs, 23 and 18, which are not the ones involved in the sex chromosome system in Alouatta palliata. The Alouatta palliata chromosomal pair 19 (X2 in males) shares homeology with chromosome 14 of Alouatta caraya.
Chromosomal homeologies between Alouatta caraya and Alouatta pigra: The chromosomal rearrangements that could explain the homeologies were grouped in two categories: 1) Alouatta pigra chromosomes with no rearrangements with respect to Alouatta caraya chromosomes: 2, 5, 6, 7, 8, 10, 15, 16, 17, 20, 22, 23, 25, 28 and X1; 2) Alouatta pigra chromosomes with more than one rearrangement with respect to Alouatta caraya chromosomes: 1, 3, 4, 9, 11, 13, 18, 19, 24, 26 and 27. No homeologies were allocated for Alouatta pigra chromosomes 4pprox, 12, 14 and 21 using the level of resolution of the classical cytogenetic techniques applied. The rearrangements detected between the Alouatta caraya and Alouatta pigra karyotypes included at least 12 fissions/fusions, two paracentric inversions, two translocations and one deletion. Alouatta caraya chromosome 7 (X2 in males) shares homeology with two Alouatta pigra chromosome pairs, 26 and 19, which are not the ones involved in the sex chromosome system in Alouatta pigra. Alouatta pigra chromosome 17 (X2 in males) shares homeology with chromosome 14 of Alouatta caraya (which in turn has homeology with HSA7).
Chromosomal homeologies among all howlers: The chromosomal homeologies found among all howlers are shown in Table 3 and Figures 1a and 1b. Results show that Mesoamerican howlers share several human chromosomal syntenic associations with South American ones: HSA15q13-q21.2/4q23-q31.2 and HSA16p/10q, shared with all howlers; HSA15q21.3-q24/14, shared with all howlers except Alouatta seniculus arctoidea and Alouatta macconnelli, and HSA8p/18, shared with all howlers except Alouatta seniculus arctoidea. Two new chromosomal syntenic associations, HSA4pter-q22/9/11 and HSA15q21.3-q24/14/21q, were found for Alouatta pigra in chromosomes 1 and 4q, respectively.
Chromosomal homeologies between howlers, obtained from data both from this contribution and from previous reports. ACA: Alouatta caraya; API: Alouatta pigra; APA: Alouatta palliata; AGU: Alouatta guariba; ASEa: Alouatta seniculus arctoidea; AMA: Alouatta macconnelli; ASA: Alouatta sara; ABE: Alouatta belzebul.
| Human Chromosomal associations |
ACA | API | APA | AGU | ASEa | AMA | ASA | ABE |
|---|---|---|---|---|---|---|---|---|
| 1p12-p21 | 1 | 3pprox | 4p | 2qter | 9qter | 18qter | 16qter | 23 |
| 5qprox-q31.2 | 13 | 3qter | 13 | 1qter | 1q | |||
| 7p22; q11; q21 | 13q | 1qter | 12qprox | 12qter | 13qter | |||
| 5q31.3-qter | 8qter | 7qter | ||||||
| 7p22; q11; q21 | 24q | 1qter | 4qter | |||||
| 6 | 2 | 2 | 3 | 2pter-qprox | 4 | 18pter-qprox | 5 | 4 |
| 8 | ||||||||
| 22q | 3 | 9 | 11 | 9p | 9qprox | 5pprox | 8qprox | 6pter |
| 9q | 1qprox | 15 | 3p | 13 | 15 | 11 | 2q | |
| 16q | 4 | 1p | 5 | 12q | 6qter | 3q | 9pter | 5 |
| 4pter-q22 | 1qprox | 11 | 14 | |||||
| 16p | 5 | 6 | 7 | 9q | 10 | 3pprox | 21 | 7 |
| 10q | 20 | 2p | 19 | |||||
| 16p | ||||||||
| 10q | ||||||||
| 8p | 6 | 5 | 6 | 4pter-qprox | 15qter | 6 | 2qter | 8 |
| 18 | 5qprox | |||||||
| 15q11-q13 | 7 (X2) | 26q | 23 | 7 (X2) | X2 | X2 | X2 | 24 |
| 15q25-qter | ||||||||
| 3p24-p21 | 19q | 18 | 17 (X2) | |||||
| 3q13-q26 | ||||||||
| 17 | 8 | 27q | 8 | 4qter | 7qprox | 7 | 1p-1qprox | 9 |
| 2p | ||||||||
| 4q31.3-qter | 9 | 8 | 10 | 11 | 18 | 10 | 16p-qprox | 11 |
| 19 | 10 | 7 | 9 | 12 | 7qter | 5pter | 15p-qprox | 10 |
| 4p | ||||||||
| 2q13-qter | 11 | 11 | 12 | 5p | 2pprox | 11qprox | 3qprox | 12 |
| 20 | 17 | 9qint | 16qprox | 8qint | ||||
| 12 | 12 | 10 | 13 | 3q | 2 | 14 | 6 | 2p |
| 11 | 13 | 1qter | 1qprox | 5q | 3qprox | 2q | 1qint | 1p |
| 12qter | 13qprox | |||||||
| 7 q11-q21 | 14 | 17 (X2) | 19 (X2) | 6 | 8 | 1q | 7 | 13 |
| 7q22-qter | ||||||||
| 3pter-p24 | 15 | 16 | 16 | 16 | 6qprox | 19 | 2qprox | 15 |
| 3p21-p12 | ||||||||
| 3q13 | ||||||||
| 3q27-qter | ||||||||
| 8q | 16 | 3q | -- | 19 | 1qprox | 12qprox | 4qprox | 16 |
| 13 | 17 | 18 | 2q | 14 | 16 | 4q | 12 | 14 |
| 2pter- q12 | 18 | 15 | 14 | 1p | 2qprox | 17 | 10 | 3q |
| 1p21-pter | 19 | 20 | 22 | 15 | 14 | 5q | 17 | 3p |
| 15q21.3-q24 | 20 | 4qint | 17 | 2qint | 6qint | 1p | 9qprox | 6pprox-qter |
| 14 | 5qter | 18 | ||||||
| 3p12 | 21 | 4pter | 1pter | 18 | 17 | 9 | 20 | 18qter |
| 21q | ||||||||
| 4q23-q31.2 | 22 | 22 | 21 | 1qint | 1pter | 1qter | 3qter | 20 |
| 15q13-q21.2 | ||||||||
| 1q32-qter | 23 | 23 | 20 | X3 | 15qprox | 21 | 8qter | 19 |
| 1q21-q31 | 24 | 25 | 25 | 8 | 3p | 16qter | 9qter | 21 |
| 10p | 25 | 28 | 24 | 22 | 7qint | 3pter | 15qter | 22 |
| X | X1 | X1 | X1 | X1 | X1 | X1 | X1 | X1 |
† from pter to qter.
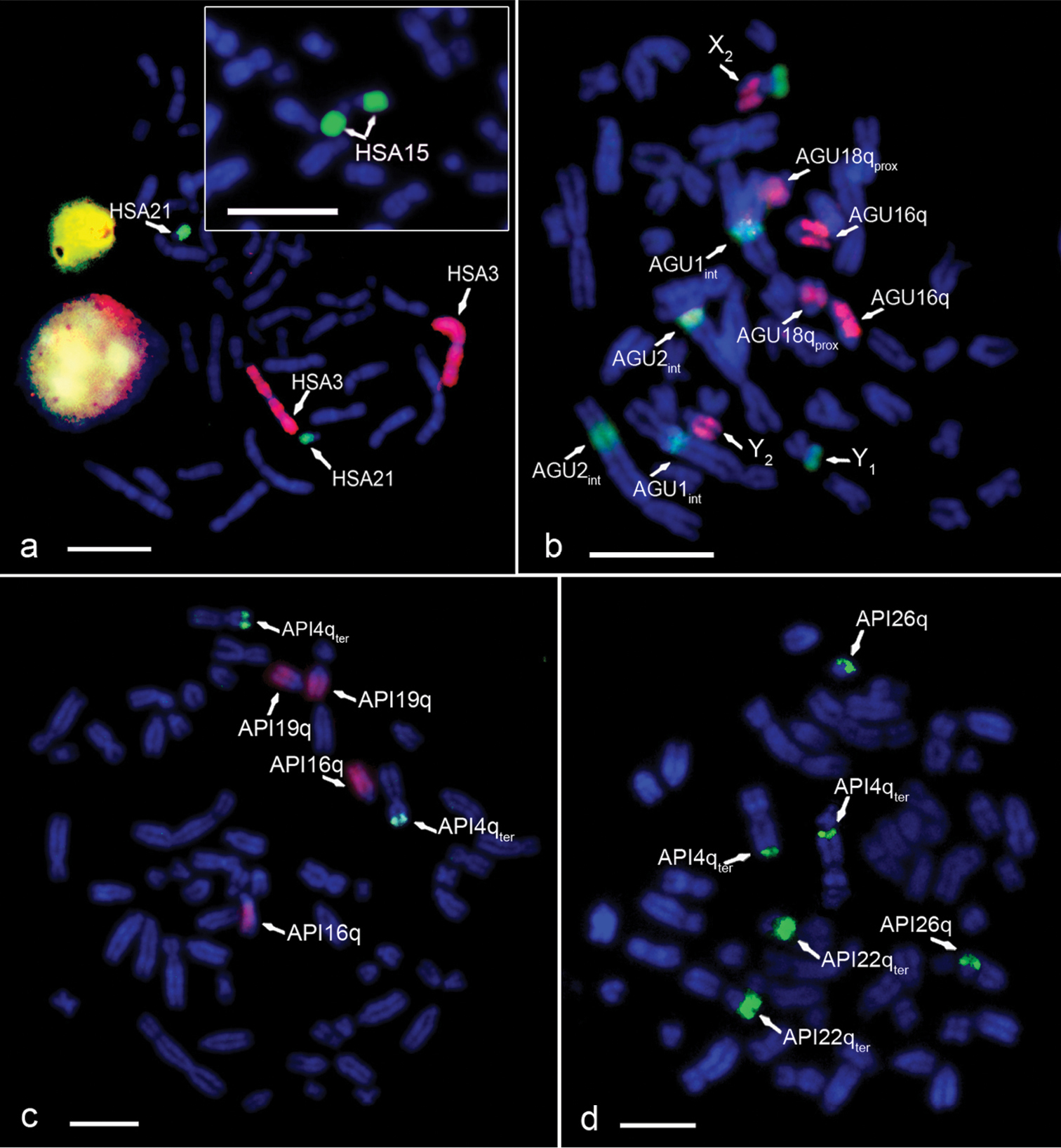
In the Homo sapiens metaphases, the hybridization signals on chromosomes HSA3, HSA21, HSA15, HSAX and HSAY for chromosome painting probes #3 (red), #21 (green), #15 (green) (Figure 2a), X and Y (data not shown) were corroborated.
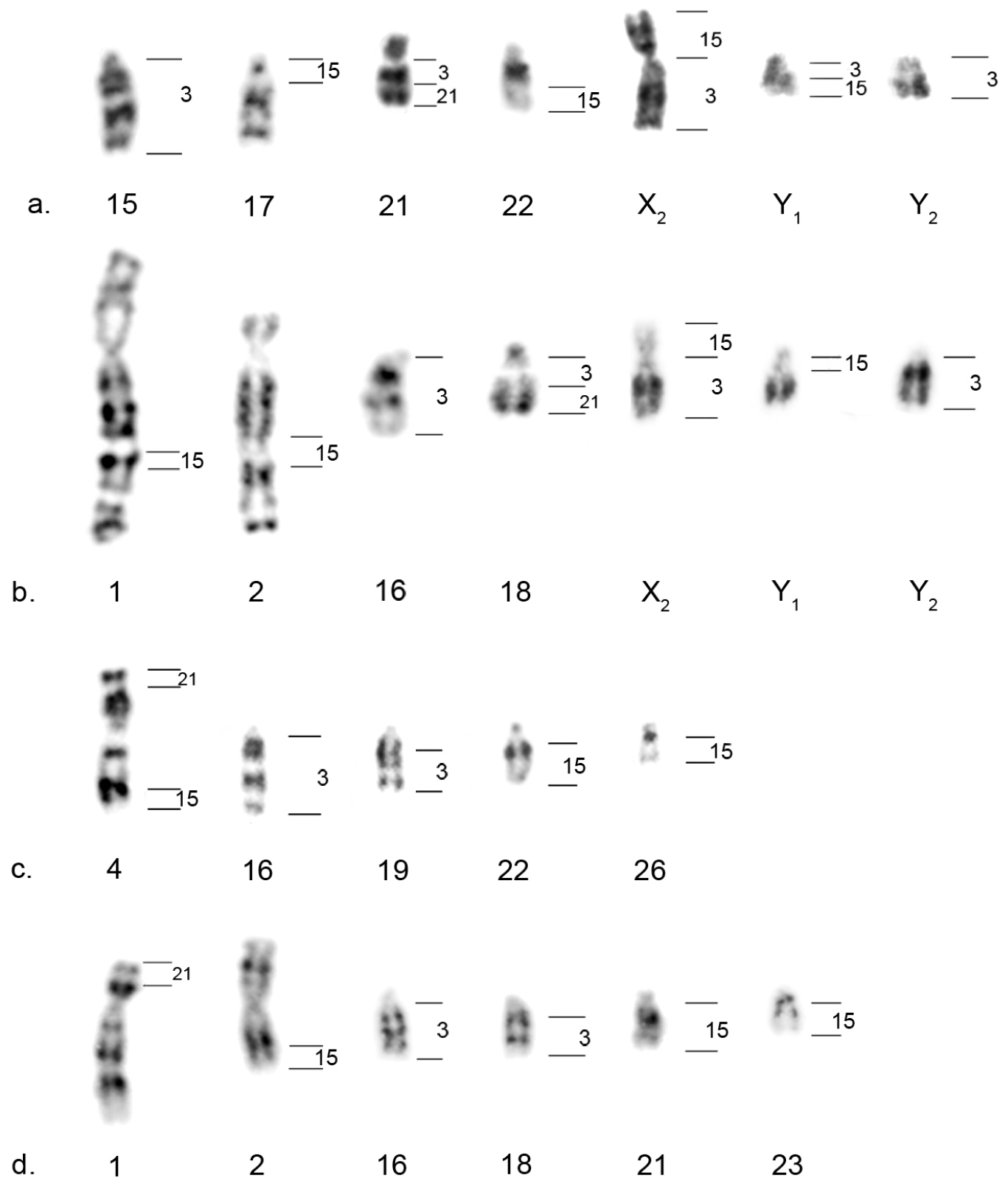
Analysis of the conservation of the HSA3/21 and HSA3/15 syntenic chromosomal associations in howlers (bar=10 μm). The arrows indicate the chromosomes with positive FISH signal a Homo sapiens partial metaphase hybridized with probes HSA21 (green) and HSA3 (red) (control of the hybridization). Inset: Homo sapiens partial metaphase hybridized with HSA15 (green) b AGUc metaphase hybridized with HSA15 (green) and HSA3 (red) c API metaphase hybridized with HSA3 (red) and HSA21 (green) d API metaphase hybridized with HSA15 (green).
In Alouatta guariba clamitans, the signal for HSA21 was observed in 18qter, the signal for HSA3 was observed in 18qprox (thus corroborating the HSA3/21 synteny in Alouatta guariba clamitans), 16q, 7q (X2 in males) and Y2, and the signal for HSA15 was observed in 1int, 2int, 7p (X2) and Y1. This corroborates the HSA3/15 syntenic association to the multiple sex chromosome system X1X1X2X2X3X3/X1X2X3Y1Y2 of this species (Figures 2b and 3b).
Howler monkeys G-banded chromosomes with positive signal for the human chromosome painting probes analyzed. On the right, the hybridization pattern of human chromosomes #3, #21 and #15. a Alouatta caraya b Alouatta guariba clamitans c Alouatta pigra d Alouatta palliata.
In Alouatta pigra, the signal for HSA3 was observed in 16q and 19q, while that for HSA21 hybridized in 4pter, thus indicating that the HSA3/21 synteny is not present in Alouatta pigra (Figures 2c and 3c). The probe for HSA15 hybridized in Alouatta pigra metaphases in 4qter, 22qter and 26q, showing that the HSA3/15 syntenic association is also absent. None of these Alouatta pigra chromosomes is involved in the sex chromosome system of this species (Figures 2d and 3c).
In Alouatta caraya, the signal for HSA21 was observed in 21qter, whereas that for HSA3 was observed in 21qprox, thus confirming the conservation of the HSA3/21 synteny. HSA15 hybridized in 7p (X2 in males) and Y1ter, and HSA3 in 7q and Y1prox, exhibiting the HSA3/15 syntenic association in the sex chromosome system X1X1X2X2/X1X2Y1Y2 (Figure 3a).
Alouatta palliata showed a pattern similar to that of Alouatta pigra (therefore Figure 2 illustrates only the latter). HSA3 hybridized in 16q and 18q, HSA21 hybridized in 1pter and HSA15 in 2qter, 21qter and 23q (Figure 3d). Both the HSA3/21 and HSA3/15 syntenic associations are absent in Alouatta palliata and chromosomes with homeology to HSA3 and HSA15 are also not involved in the sex chromosome system of this species.
The probe for the human X chromosome showed positive hybridization signal in X1 of all the species analyzed. The probe for the human Y chromosome did not hybridize in any of the howler species (data not shown).
The data obtained from the G-banding pattern and FISH homeologies, together with cyt b sequences obtained from previous reports, were used as the basis to perform a cladistic analysis. The HSAY/7 association, corresponding to the Y-autosome translocation that gave rise to the multivalents observed in Alouatta pigra and Alouatta palliata, was added as an extra character to the original list (
Three data matrices were obtained: one including only chromosomal data, another including only molecular data and the last one including both types of characters (chromosomal and molecular) in a single matrix (see Appendix 1).
Chromosomal partition: The analysis of chromosomal data resulted in 36 informative characters, 23 constant characters and 40 non-informative characters. After analyzing 704 trees, PAUP retained the two most parsimonious trees (Appendix 2: Figures Sa and Sb), both with a length of 87 (L = 87). The analysis using only the partition of chromosomal data did not resolve the node ((Alouatta palliata, Alouatta pigra), (Alouatta caraya, Alouatta belzebul), ((Alouatta guariba clamitans, Alouatta guariba guariba), (Alouatta macconnelli (Alouatta sara, Alouatta seniculus arctoidea))), since it was established in a polytomy (Appendix 2: Figure Sc).
Molecular partition: Heuristic analysis of cyt b gene sequences, made from a total of 800 characters, produced 109 informative characters, 551 constant characters and 140 non-informative characters. After analyzing 916 trees, PAUP retained a single most parsimonious tree (Appendix 2: Figure Sd), with a length of L = 366. The analysis using only molecular data did not resolve the node (Alouatta sara, Alouatta macconnelli, Alouatta seniculus arctoidea, Alouatta caraya), which was established as a polytomy different from that described from chromosomal data.
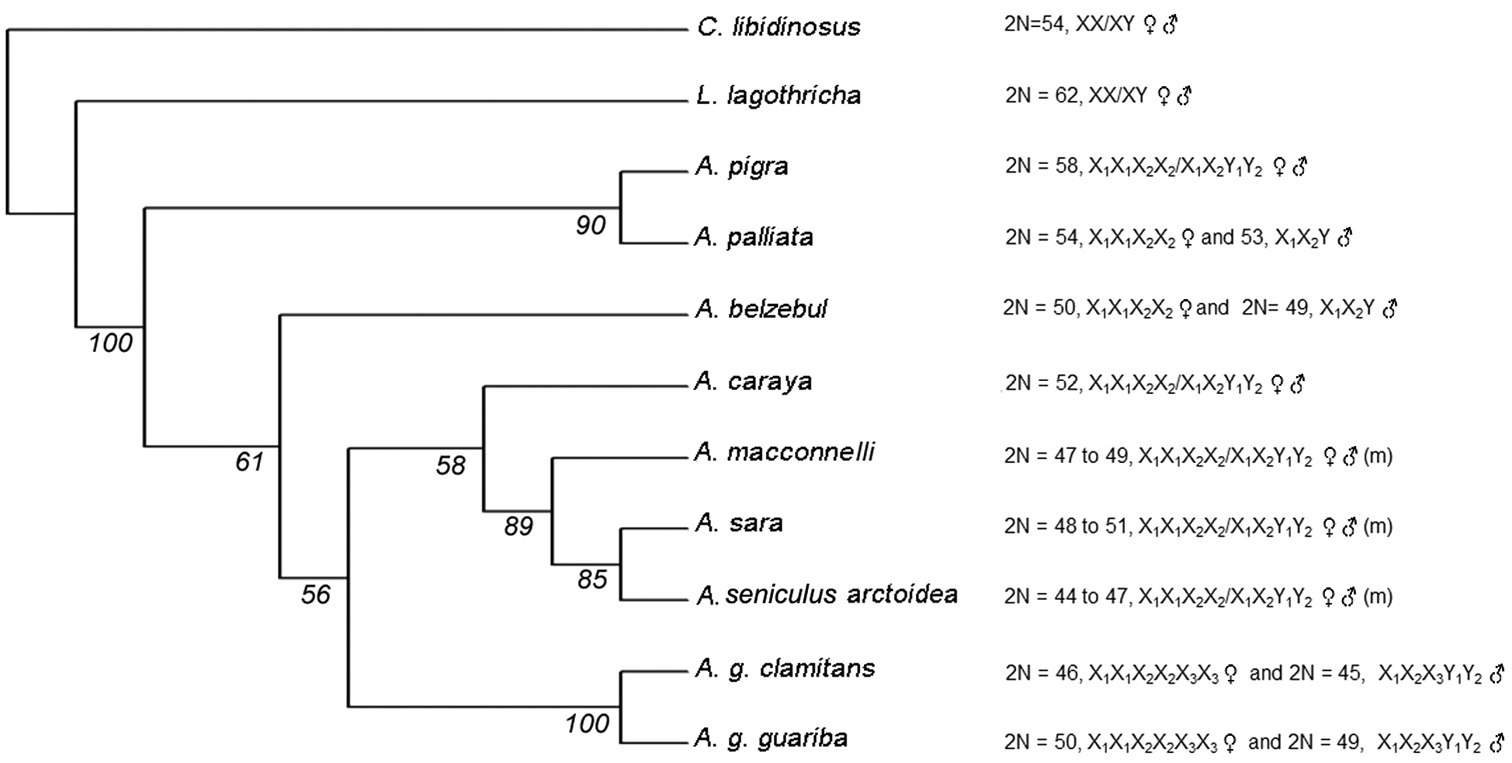
Combined analysis: The heuristic analysis of the combined data showed a total of 899 characters, 145 of which were informative, 180 non-informative and 574 constant. After analyzing 684 trees, PAUP retained only one, with a length of L=460 (Figure 4). This type of analysis allowed us to solve all the nodes, resulting in a fully resolved tree.
50% majority consensus tree obtained by “bootstrap” for the combined analysis. Next to the name of each species, the diploid number (2N) and sex chromosome system is described. (m)=microchromosomes.
We present the first phylogenetic study using a combined analysis of chromosomal and molecular characters in Ceboidea to contribute to the characterization of the speciogenic processes in howler monkeys. The homoplasy distribution is likely to be different in each dataset because these are subject to different constraints. Therefore, when different datasets are analyzed simultaneously, the signal common to all of them is more likely to overwhelm the homoplasy signal on the data (
In primates, few studies have compared and taken into account more than one type of character.
In all the above-mentioned contributions, the X1X1X2X2/X1X2Y1Y2 sex chromosome system was proposed as the ancestral condition for the genus. However, as discussed by
The karyotypes of Alouatta pigra and Alouatta palliata share more syntenic associations with those of Alouatta caraya and Alouatta belzebul than with those of the “Alouatta seniculus group” (Alouatta seniculus arctoidea, Alouatta sara, Alouatta macconnelli, denominated as such because they were once all subspecies of Alouatta seniculus together with Alouatta seniculus seniculus Linnaeus, 1766, and Alouatta seniculus stramineus Hill, 1962). This supports the basal grouping of the Alouatta pigra-Alouatta palliata Mesoamerican clade and the basal grouping of Alouatta belzebul among South American howlers.
The chromosomal comparisons showed that Alouatta pigra and Alouatta palliata conserved the HSA8/18 and HSA14/15 syntenies, considered ancestral for Platyrrhini (
According to our combined phylogeny, the HSA2/20 and HSA5/7/5/7 syntenic associations, previously considered as synapomorphies of the Alouatta caraya-Alouatta belzebul group (
Like the HSA3/21 synteny, the HSA3/15 syntenic association, involved in the sex chromosome systems in South American howlers, is not present in Mesoamerican ones. This syntenic association of human 3/15 chromosomal segments has been described in other Atelidae species such as Ateles geoffroyi Kuhl, 1820 and Ateles belzebul hibridus Geoffroy, 1829, although not associated with the sex chromosome system (
Taking into consideration the data obtained, a hypothesis can be proposed regarding the origin of the sex chromosome systems in the genus. Within the family Atelidae, with the exception of Alouatta, all genera have an XX/XY sex chromosome system. Therefore, it can be considered that the Alouatta ancestor possessed a chromosomal sex determination XX/XY, prior to the biogeographic separation of Mesoamerican and South American groups (see below). After this separation, both groups independently acquired the multiple sex chromosome systems currently observed through independent Y-autosome translocations.
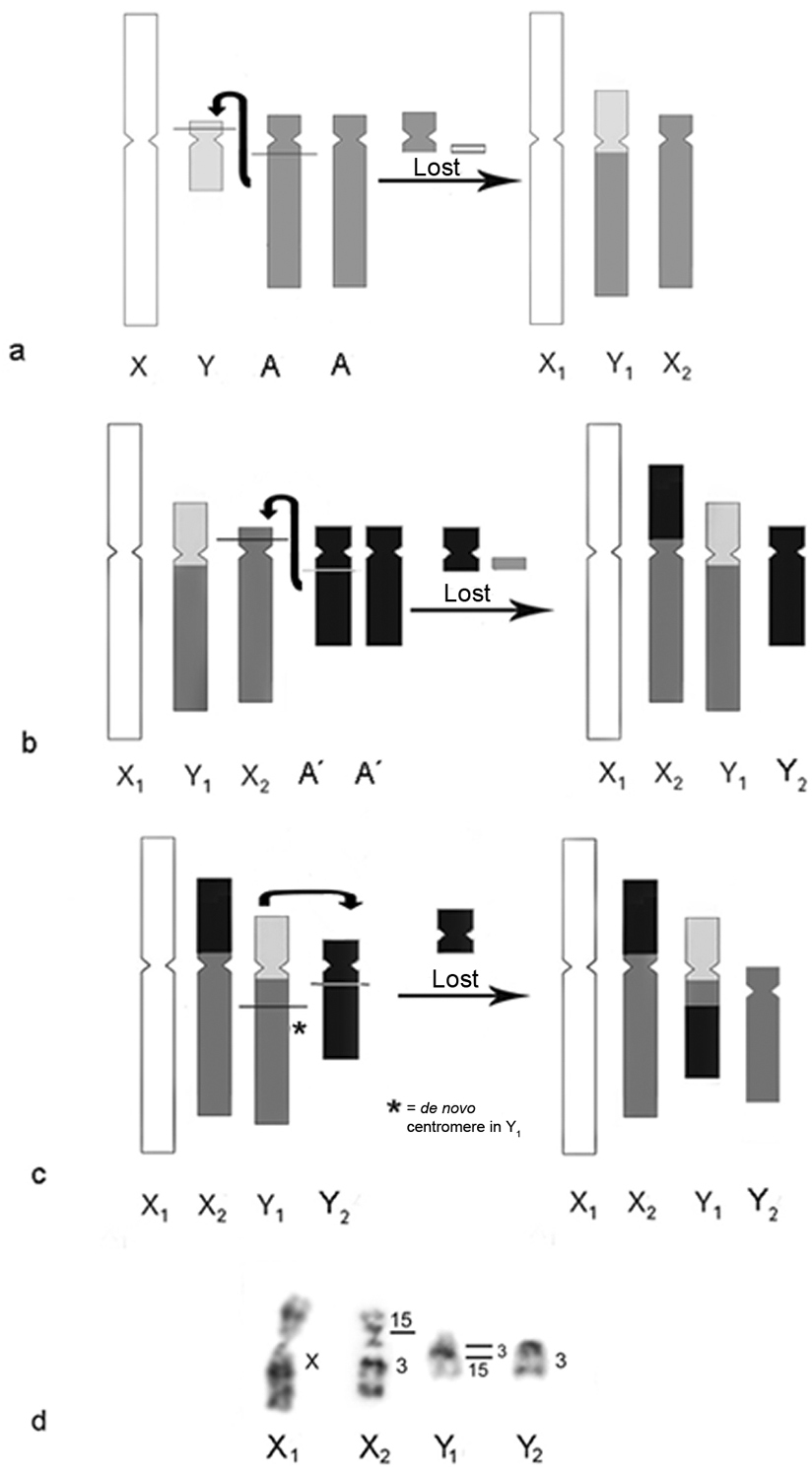
The sex chromosome system X1X1X2X2/X1X2Y would have arisen independently in the lineages of Meso and South American howlers by a Y-autosome translocation (Figure 5a). In males, two fissions, one in Ypter and another in qprox of the autosomal pair involved (Aqprox), followed by translocation of Aqprox to Yq-pprox, formed the new chromosome Y1. The Ypter segment is lost and the proximal region of the fissioned autosome either is lost or, in certain howler species, could have given rise to microchromosomes (e.g.: Alouatta seniculus (
a Possible origin for X1X1X2X2/X1X2Y sex chromosome system in the genus Alouatta. The ancestral X chromosome is shown in white, the ancestral Y chromosome in light gray and the autosomal pair (A) in dark gray. Two fissions occurs, one in Ypter and another in qprox of the autosomal pair involved (Aqprox). The translocation of Yq-pprox to the Aq formed the new Y1 chromosome and the homolog of the autosomal pair involved in the translocation is now denominated X2. The Ypter acentric fragment is lost and the rest of the autosome (Ap and Aqprox) could either be lost or remain as a microchromosome in some howlers b Possible origin for the X1X1X2X2/X1X2Y1Y2 sex chromosome systems from a X1X1X2X2/X1X2Y system. The ancestral X is shown in white, the ancestral Y in light gray, the autosomal pair involved in the first translocation (A) in dark gray and the autosomal pair (A´) involved in the formation of this new sex chromosome system in black. Simultaneous breaks in X2pprox and A`qprox followed by the translocation of the rest of the A`q to X2pprox give origin to the new X2 chromosome. The X2pter acentric fragment could be lost and the rest of the autosome (A´) could either be lost or remain as a microchromosome in some howlers. The homolog to the autosomal chromosome in question is now identified as Y2c Simultaneous breaks in Y1q and Y2q and a translocation between Y1 and Y2 further explain the hybridization pattern observed in the sex chromosome systems of South American howlers. A de novo centromere arises in the remains of the old Y1 (now Y2). The remains of the old Y2 could either be lost or remain as a microchromosome in some howlers d Hybridization pattern in South American howlers.
From this X1X1X2X2/X1X2Y sex chromosome system, an X1X1X2X2/X1X2Y1Y2 system could have arisen from a new translocation (Figure 5b). Under this hypothesis, simultaneous breaks in X2pprox and qprox of another autosome (A´qprox), followed by the translocation of most of the A´q arm to X2pprox, gave rise to the new X2 chromosome. The X2pprox acentric fragment is lost and the rest of the autosome (A´) either is lost or could have remained as a microchromosome in some howler species (see above). The chromosome homologous to the autosome in question (A`) became Y2. In the case of South American howlers, the new autosomal pair involved in the sex chromosome system would share homeology with HSA15. A further translocation between Y1 and Y2 (Figure 5c) would explain the hybridization pattern of the segments with homeology to human chromosomes 3 and 15 observed in the sex chromosome systems X1X1X2X2/X1X2Y1Y2 in South American howlers (Figure 5d).
On the other hand, in the Mesoamerican species, the X1X1X2X2/X1X2Y1Y2 sex chromosome system could have arisen either as described in Figure 5b (with the autosomal pair involved sharing homeology with a human chromosome not yet identified by G-banding pattern) or by a fission in Y1 that would have given rise to two chromosomes, the new Y1 (containing the segment corresponding to the ancestral Y chromosome) and Y2 (containing a portion of the autosomal pair with homeology to HSA7). This last hypothesis would require a centromeric activation in Y2.
However, considering the observation of the independent origin of the multiple sex chromosome systems in these two groups of howlers, the possibility of an independent origin of the X1X1X2X2/X1X2Y and X1X1X2X2/X1X2Y1Y2 sex chromosome systems within the Meso and South American groups cannot be ruled out until further studies.
It can be considered that multiple sex chromosome systems would be an extremely rare phenomenon due to complication in meiosis. Extreme cases are platypus and echidna, with a large number of sex chromosomes (
The chromosomal homeologies and FISH analysis were used to construct a data matrix for the phylogenetic analysis. For comparison purposes, independent phylogenetic reconstructions were performed with each type of partition (Appendix 2: Figure Sa, b, c and d), along with the combined analysis of the two datasets (Figure 4). The chromosome partition grouped Alouatta caraya and Alouatta belzebul as sister taxa, in agreement with that reported by
The grouping of South American species as a separate group of the Mesoamerican group coincides with previous phylogenetic analyses using only molecular characters (
Independently of the biogeographic scenario under consideration, it is clear that the evolutionary history of Mesoamerican howlers is different from that of South American howlers, an assertion that would be supported by the evidence provided by our new data.
This contribution provides new useful information for the systematics of the genus Alouatta, while supporting the hypothesis of chromosomal evolution in primates as a speciogenic strategy. The combined analysis resolved the phylogenetic relationships between howler species of both American origins, as a first approach to the “Total Evidence” concept and towards clarifying the controversies related to the Taxonomy and Evolution of Ceboidea.
All research reported in this manuscript has met the appropriate national and institutional guidelines for the legal acquisition and use of laboratory animals and authorized study of wild animals. The authors also adhered to the Guide for Care and Use of Experimental Animals, as promulgated by the Canadian Council of Animal Care, and to the American Society of Primatologists (ASP) Principles for the Ethical Treatment of non-human Primates. The work which took place both in Argentina as in Mexico was done in accordance with the laws of these countries.
We would like to thank the head of the institutions that made possible the sampling of the specimens for the genetic studies presented here, the veterinarians who collaborated in the sampling of all specimens, and the caretakers of the animals. To Dr. Liliana Cortés-Ortiz (Dept. of Ecology and Evolutionary Biology & Museum of Zoology, University of Michigan), Dr. Domingo Canales-Espinosa; Vet. Javier Hermida, Sr. Francisco García-Orduña (Instituto de Neuroetología, Universidad Veracruzana, Xalapa, Veracruz, México) and all the field team for their collaboration and support in the obtention of the samples of mexican howlers in their natural geographic distribution. MDM PIP-CONICET 0744 and UBACyT X154 provided funding for this project.
Data matrix. (doi: 10.3897/CompCytogen.v8i1.6716.app1) File format: Microsoft Word file (doc).
Explanation note: Data matrix contains: Chromosomal data matrix, Molecular data matrix and Combined data matrix.
Supplementary Figure S. (doi: 10.3897/CompCytogen.v8i1.6716.app2) File format: Microsoft Word file (doc).
Explanation note: Figure S: a) and b) Most parsimonious trees obtained for the chromosomal partition c) 50% majority consensus tree obtained by “bootstrap” d) 50% majority consensus tree obtained by “bootstrap” for the molecular partition.