(C) 2014 Camilla Bruno Di-Nizo. This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Di-Nizo CB, Neves CL, Vilela JF, Silva MJJ (2014) New karyologycal data and cytotaxonomic considerations on small mammals from Santa Virgínia (Parque Estadual da Serra do Mar, Atlantic Forest, Brazil). Comparative Cytogenetics 8(1): 11–30. doi: 10.3897/CompCytogen.v8i1.6430
Atlantic Forest, in the eastern coast of Brazil, is a hotspot of biodiversity of mammals, and Parque Estadual da Serra do Mar (PESM) is the largest continuous area of this biome. Here, we characterized the karyotype composition of the small mammals from Santa Virgínia, a region in the northern part of PESM. Specimens were collected from July 2008 to September 2009. We identified 17 species (13 rodents and 4 marsupials) from which 7 exhibited species-specific karyotypes, illustrating the importance of karyotype information in cytotaxonomy. We report for first time the karyotype of Monodelphis scalops (Thomas, 1888) and two new records for PESM: Akodon montensis Thomas, 1913 and Brucepattersonius soricinus Hershkovitz, 1998. Cytogenetic polymorphisms were detected for some species trapped in the area. Our results show the importance of Santa Virgínia / PESM in addressing studies for the conservation of small mammal wildlife in the Atlantic Forest.
Atlantic Forest, conservation, cytotaxonomy, Monodelphis scalops
The Atlantic Forest is the fourth biodiversity hotspot in the world (
The Parque Estadual da Serra do Mar (PESM), located in the state of São Paulo, Brazil was created in 1977, and is considered the largest remaining block of Atlantic Forest with 315.390 hectares (
Studies the mammal fauna of this park are scarce and the majority of the reports were presented in undergraduate theses and master’s dissertations, focusing on large mammals (
According to
This study aims to characterize the karyotype composition and contribute to the identification of small rodents and marsupials from Santa Virgínia, since there is only one published study focusing on small mammals of this area. Data about geographical distribution of trapped species are also given.
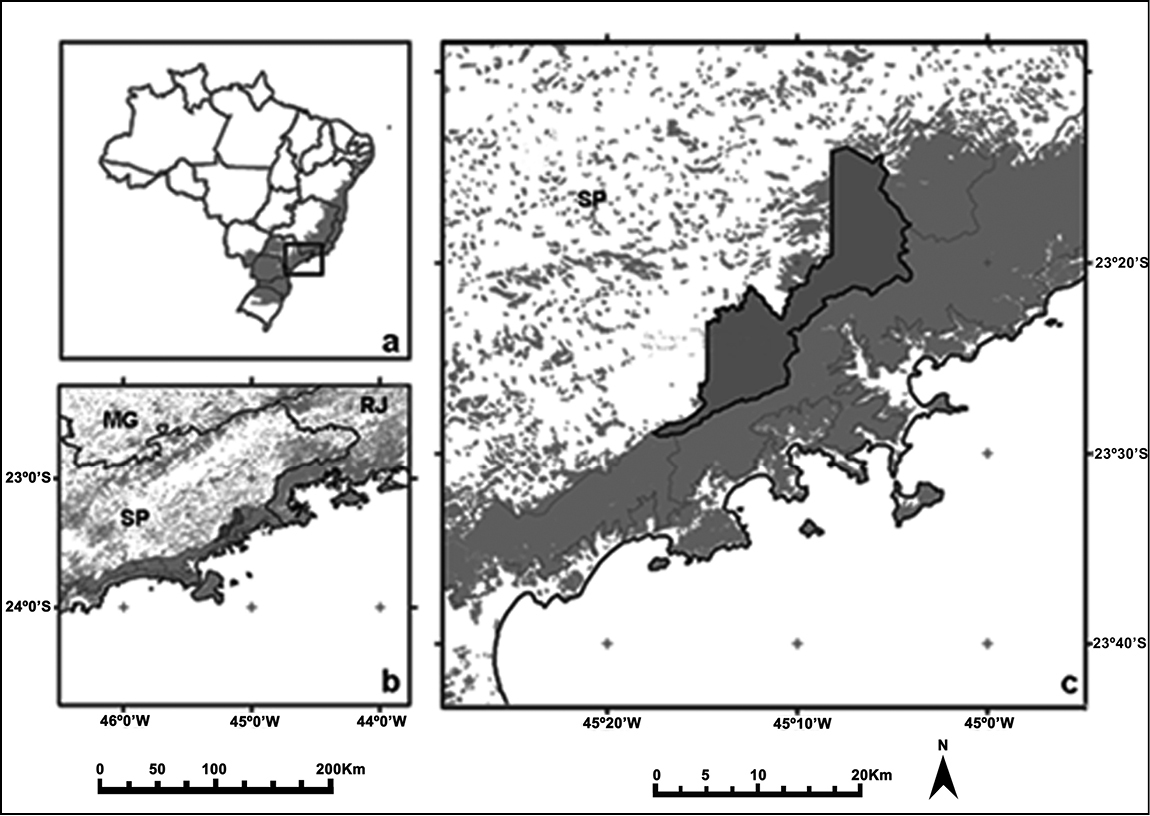
Santa Virgínia (lat. 23°24.00'S to 23°17.00'S, long. 45°03.00'W to 45°11.00'W) is located in the Northern of PESM (Fig. 1) covering an area of 17, 000 hectares (
a Map of Brazil with original Atlantic Forest cover in grey and the region of Parque Estadual da Serra do Mar (PESM) indicated (square) b Parque Estadual da Serra do Mar (PESM) in grey c Santa Virgínia is highlighted (extracted and modified from
Small mammals were sampled by commercial live-traps (Sherman and Tomahawk-like traps) and pitfall-traps. In July 2008, a pilot experiment was performed from one to three nights, with a total sampling effort of 300 live-traps/night. From September 2008 to September 2009, field survey was carried out bimonthly during five consecutive nights. During this period, we set up six grids with 30 live-traps per grid and 12 transects of pitfall-traps. Live-traps were arranged in a 0.6 ha grids (60 × 100 m each) with 24 trap stations spaced every 20 meters. Each trap station received one Sherman of different size, randomly set (small, 25 × 7.5 × 9.5 cm; medium, 30 × 7.5 × 9.5 cm; large, 37.5 × 10 × 12 cm; H.B. Sherman Trap®, Inc., Tallahassee, Florida, USA). We also set randomly a Tomahawk-like trap (45 × 16 × 16 cm; Rosaminas Serviço Engenharia e Comércio Ltda. Piraúba, Minas Gerais, Brazil) at six trapping stations. Overall, we had 6300 live-trap/night.
The 12 transects of pitfall-traps were pairwise 30 meters apart, from November 2008 to September 2009. Each transect received four plastic buckets (60L, 40 cm top diameter, 35 cm bottom diameter, and 56 cm depth) buried with the rim at ground level, spaced every 10 meters each. The buckets on each line were connected with a 0.5 meters tall plastic drift fence that extended an additional 10 meters at each end, totaling 50 meters of fence. In total, we used 48 buckets, resulting in 1, 440 pitfall-traps/night.
Different sizes and models of traps were used to optimize the sampling, aiming to reduce the selectivity based on body size and/or habits of the animals. Attractive baits (mashed bananas, peanut butter, bacon and corn meal) were placed in both kinds of traps. All traps were checked daily, preferably on the first hours in the morning.
Trapping and handling were carried out under ICMBio licence (number 14428-2) of Instituto Chico Mendes de Conservação da Biodiversidade.
Animals were euthanized according to the protocol of the “Animal experimentation ethics” (
A list of cytogenetically studied small mammals from Santa Virgínia, Parque Estadual da Serra do Mar, state of São Paulo, Brazil. N: number of individuals analyzed. Specimens voucher/Museum Field number: ROD and MARS - Laboratório de Ecologia e Evolução Instituto Butantan, Brazil; MN - Museu Nacional, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil; UFES - Coleção de Mamíferos da Universidade Federal do Espírito Santo, Brazil. 2n: diploid number, and FNa: number of autosomes arms. Morphologies: A=acrocentric; M=metacentric; SM=submetacentric; ST=subtelocentric. Grey cells correspond to species-specific karyotypes.
|
ORDER Family Tribe Species |
N | Specimens voucher/ museum field number | Distribution | 2n | FNa | Autosome pairsª | Sex chromosomes | Variable cytogenetic characteristics | Karyotype reference | Figure No. |
|---|---|---|---|---|---|---|---|---|---|---|
|
ORDER RODENTIA Family Cricetidae Tribe Akodontini Akodon montensis |
3♀6♂ | ROD 3*, 6*, 11*, 28*, 29* UFES 2235-2237, 2239 | From Rio de Janeiro to Rio Grande do Sul and Minas Gerais, Brazil 1, 2 | 24, 24 (+ 1B) | 42 | 9 large to medium M/SM; 1 A; 1 small M | X: medium A Y: small A |
X chromosome polymorphism (enlarged short arm), 1 SM B-chromosome |
|
3a |
| Blarinomys breviceps | 1♀ | UFES 2263 | Endemic of Atlantic Forest, Brazil1, 2 | 29 (+2B) | 50 | 11 medium M/SM 1 A Heteromorphic pair:1 M + 2 A |
X: large A | Heteromorphic pair, 2 M B-chromosomes |
|
See |
| Brucepattersonius soricinus | 1♀, 1♂ | MN 78955, 78956 | Southeastern Brazil, exclusively in Atlantic Forest1, 2, 3 | 52 | 52 | 24 medium to small A; 1 small SM | X: large ST Y: small A |
- |
|
3b–d |
| Thaptomys nigrita | 2♂ | ROD 2*, 4* | South Bahia to the north of Rio Grande do Sul, Brazil1, 2 | 52 | 52 | 24 medium to small A; 1 small SM |
X: large A Y: small SM |
- |
|
5a |
| Tribe Oryzomyini Drymoreomys albimaculatus |
1♀, 1♂ | UFES 2271, 2272 | Endemic of Atlantic Forest, Brazil 4 | 62 | 62 | 29 medium to small A; 1 small M | X: large SM Y large SM, smaller than the X |
- |
|
See |
| Euryoryzomys russatus | 1♀, 7♂ | ROD 5*, 12*, 30* UFES 2242- 2244, 2265-2266 |
Coastal region of Brazil from Bahia to Rio Grande do Sul1, 2 | 80 | 86 | 35 A decreasing in size; 4 small M | X: large SM Y: small A or small ST |
Sex chromosomes polymorphisms |
|
5b |
| Nectomys squamipes | 1♀ | UFES 2270 | Eastern Brazil 2 | 56 (+2B) | 56 | 26 A decreasing in size; 1 small M | X: large SM | 2 small SM B-chromosomes |
|
4a |
| Oligoryzomys nigripes | 4♀, 4♂ | ROD 34*, UFES 2274-2280 | From South Bahia to Rio Grande do Sul, Brazil1, 2 | 62 | 80–82 | 11 M/SM decreasing in size; 19 A decreasing in size | X: large SM or large M Y: medium M or medium SM |
Pericentric inversions in pair 3, sex chromosomes polymorphisms |
|
4b |
| Sooretamys angouya | 1♀, 4♂ | UFES 2262, 2282-2285 | From Espírito Santo to Santa Catarina, Brazil2 | 58 | 60 | 26 A decreasing in size; 2 small M | X: large A Y: medium A |
- |
|
5c |
| Tribe Phyllotini Calomys tener |
1♂ | UFES 2264 | Widespread in the state of São Paulo, Brazil1, 2 | 66 | 66 | 31 medium to small A; 1 M | X: large SM Y: medium A |
- |
|
6a |
| Tribe Thomasomyini Rhipidomys itoan |
1♀ | UFES 2281 | PESM5, 6 | 44 | 50 | 17 A decreasing in size; 1 medium SM; 3 small M | X: large SM | - |
|
4c |
|
Incertae sedis Juliomys pictipes |
3♂ | UFES 2267-2269 | Minas Gerais to Rio Grande do Sul, Brazil1, 2 | 36 | 34 | 17 A decreasing in size | X: medium A Y: small A |
- |
|
6b–c |
|
Family Echimyidae Trinomys iheringi |
2♀, 1♂ | ROD 7*, 10*, UFES 2286 | West of Rio de Janeiro, São Paulo to north of Paraná, Brazil2, 7 | 60+1B, 60+4B | 116 | 29 M or SM decreasing in size | X: large SM Y: small SM |
1 or 4 dot-like B-chromosomes; Secondary constriction on pair 7 |
|
4d |
|
ORDER DIDELPHIMORPHIA Family Didelphidae Marmosops incanus |
2♀, 1♂ | MARS 1*, 5*, 6* | Eastern Brazil8 | 14 | 24 | 6 SM decreasing in size | X: small SM Y: small A |
- |
|
7a |
| Micoureus paraguayanus | 1♀, 1♂ | MARS 3*, 4* | Atlantic Forest; Eastern Brazil, until Rio Grande do Sul state8 | 14 | 20 | 4 M or SM 2 A |
X: medium A Y: medium A, smaller than X |
- | Pereira et al. (2008) | 7b |
| Monodelphis scalops | 1♂ | MN 78961 | Espírito Santo, Rio de Janeiro and São Paulo, Brazil8 | 18 | 30 | 4 SM 3 ST 1 A |
X: small ST Y: minute A |
- | Present study | 2 |
| Philander frenatus | 1♀, 1♂ | UFES 2287-2288 | From Bahia to Santa Catarina, Brazil8 | 22 | 20 | 10 A | X: medium A Y: small A |
|
7c |
Geographic distribution according to: 1.
*Specimens voucher deposited in Museu de Zoologia da Universidade de São Paulo (MZUSP) without catalog number yet. ª Autosomal morphologies do not include Bs.
The nomenclature used in this work follows
Metaphases were obtained from bone marrow and spleen after in vivo injection of a 0.1% colchicine solution (1mL/100g of weight). Cells were suspended in 0.075M KCl solution for 20 minutes at 37°C and fixed in three washes of methanol: acetic acid (3:1). GTG and CBG-banding were performed according to
Specimen identification was carried out through a comparison of our data with previous cytogenetic information, external morphological characteristics, and geographic distribution (see Table 1 references).
A total of 706 small mammal specimens were captured (600 rodents and 106 marsupials) and 54 specimens were selected for chromosome preparations (46 rodents and 8 marsupials, Table 1).
On the whole, 13 species of rodents belonging to two families were cytogenetically analyzed (Table 1): Akodon montensis Thomas, 1913; Blarinomys breviceps (Winge, 1887); Brucepattersonius soricinus Hershkovitz, 1998; Thaptomys nigrita (Lichtenstein, 1829); Drymoreomys albimaculatus Percequillo, Weksler & Costa, 2011; Euryoryzomys russatus (Wagner, 1848); Nectomys squamipes (Brants, 1827); Oligoryzomys nigripes (Olfers, 1818); Sooretamys angouya (Fischer, 1814); Calomys tener (Winge, 1887); Rhipidomys itoan Costa, Geise, Pereira and Costa, 2011; Juliomys pictipes (Osgood, 1933) of family Cricetidae, and Trinomys iheringi (Thomas, 1911) of family Echimyidae.
Four marsupial species (Didelphimorphia) were karyotyped: Marmosops incanus (Lund, 1840); Micoureus paraguayanus (Tate, 1931); Monodelphis scalops (Thomas, 1888) and Philander frenatus (Olfers, 1818) (Table 1).
Eight individuals were collected, although only one male had been cytogenetically studied. Morphological data and geographic distribution comparisons allow us to identify all as Monodelphis scalops. The morphological traits of these individuals are similar to voucher specimens of Monodelphis scalops preserved at MZUSP under catalogue numbers 1528, 30702, 30712 and 30757. This species has also been reported in São Paulo state, Brazil (
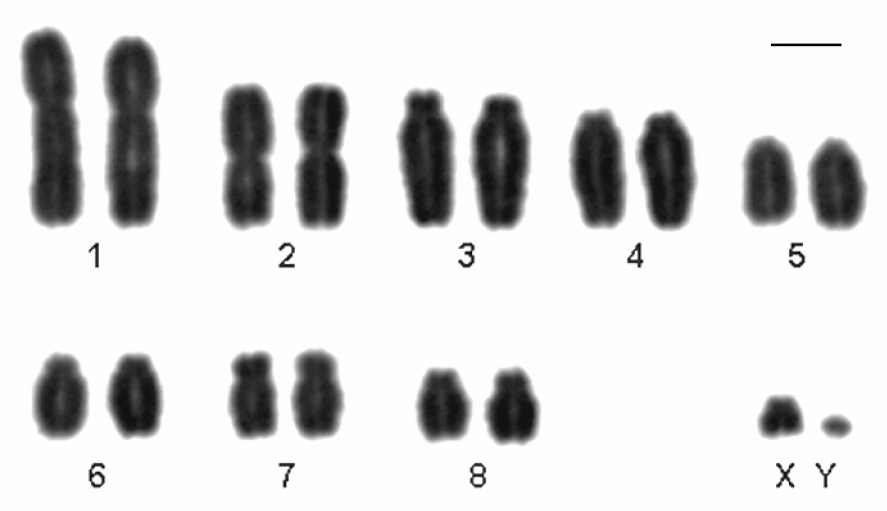
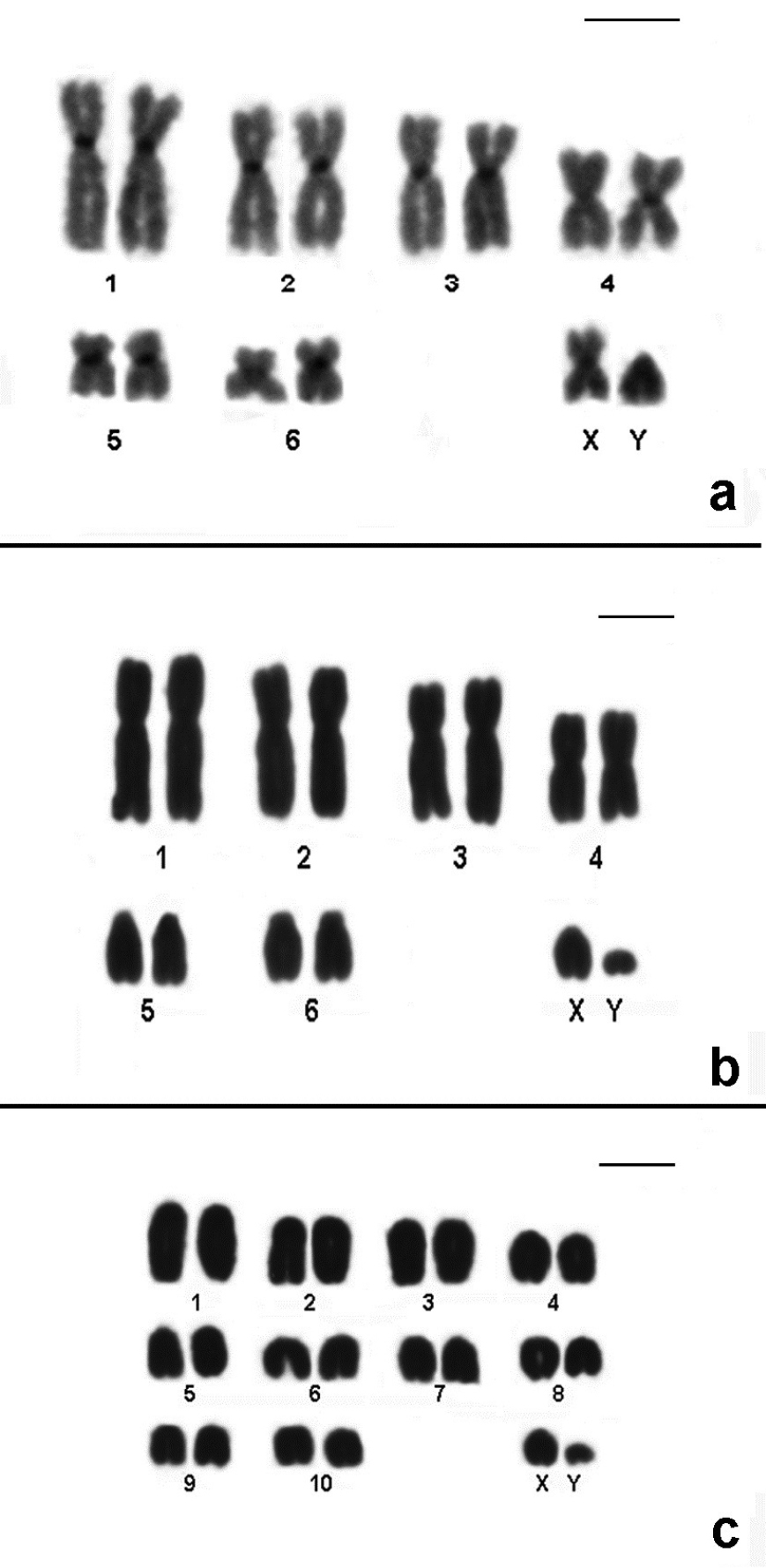
Here we present, for the first time, the karyotype of Monodelphis scalops. The karyotype of a male showed 2n=18, FNa=30. Pair 1 is a large submetacentric, pair 2 is a medium metacentric, pairs 3, 4 and 6 are medium subtelocentric, pair 5 is a medium acrocentric and pairs 7 and 8 are medium submetacentric. X chromosome is a small subtelocentric, and the Y is a minute acrocentric (Fig. 2). The short arm of pairs 4 and 6 are difficult to see depending on the condensation of the chromosome and so it was necessary to analyze and measure more than 30 metaphases to define their morphology.
Conventional stained karyotype of Monodelphis scalops (2n=18, FNa=30, male). Bar = 10µm.
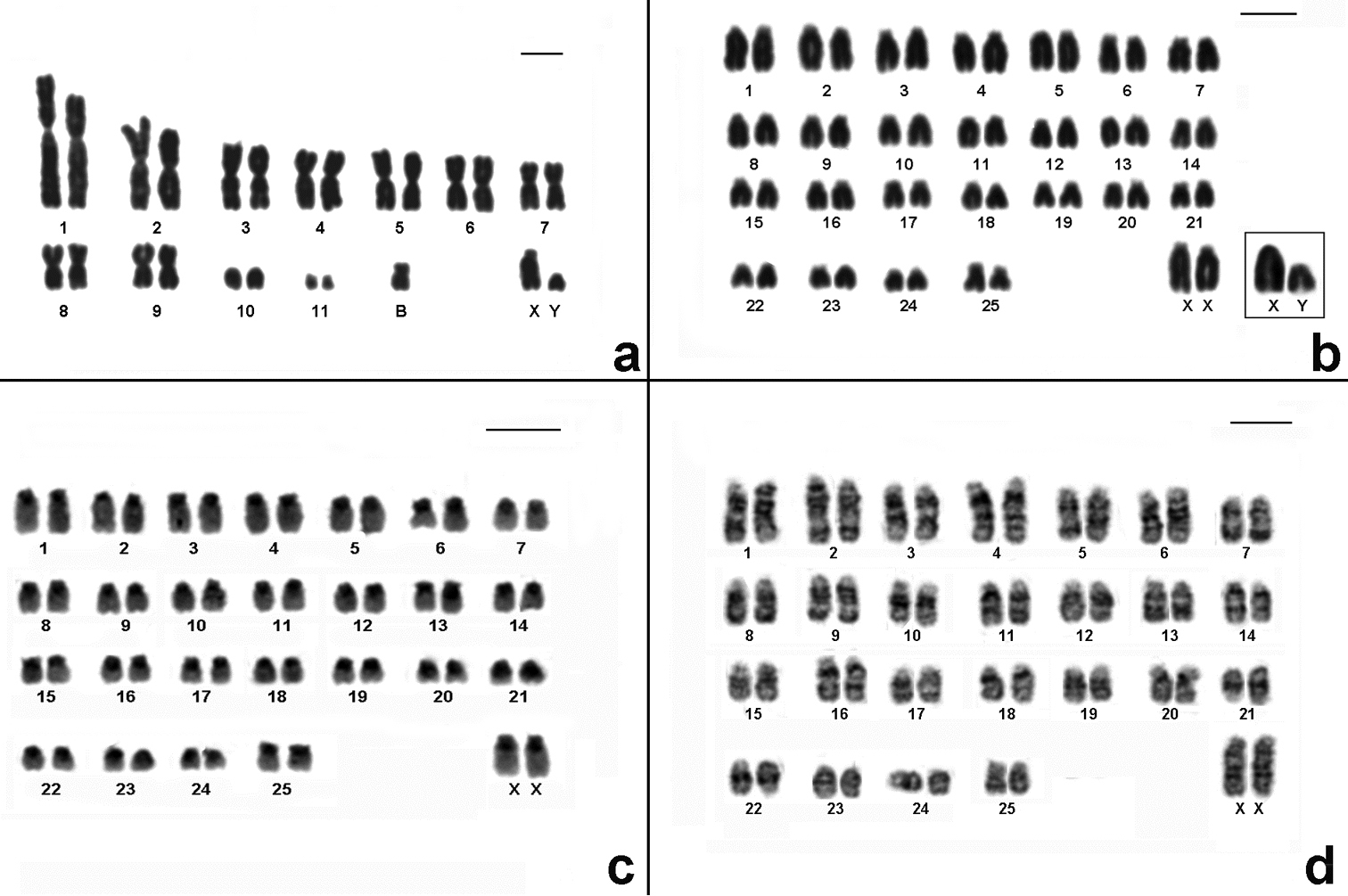
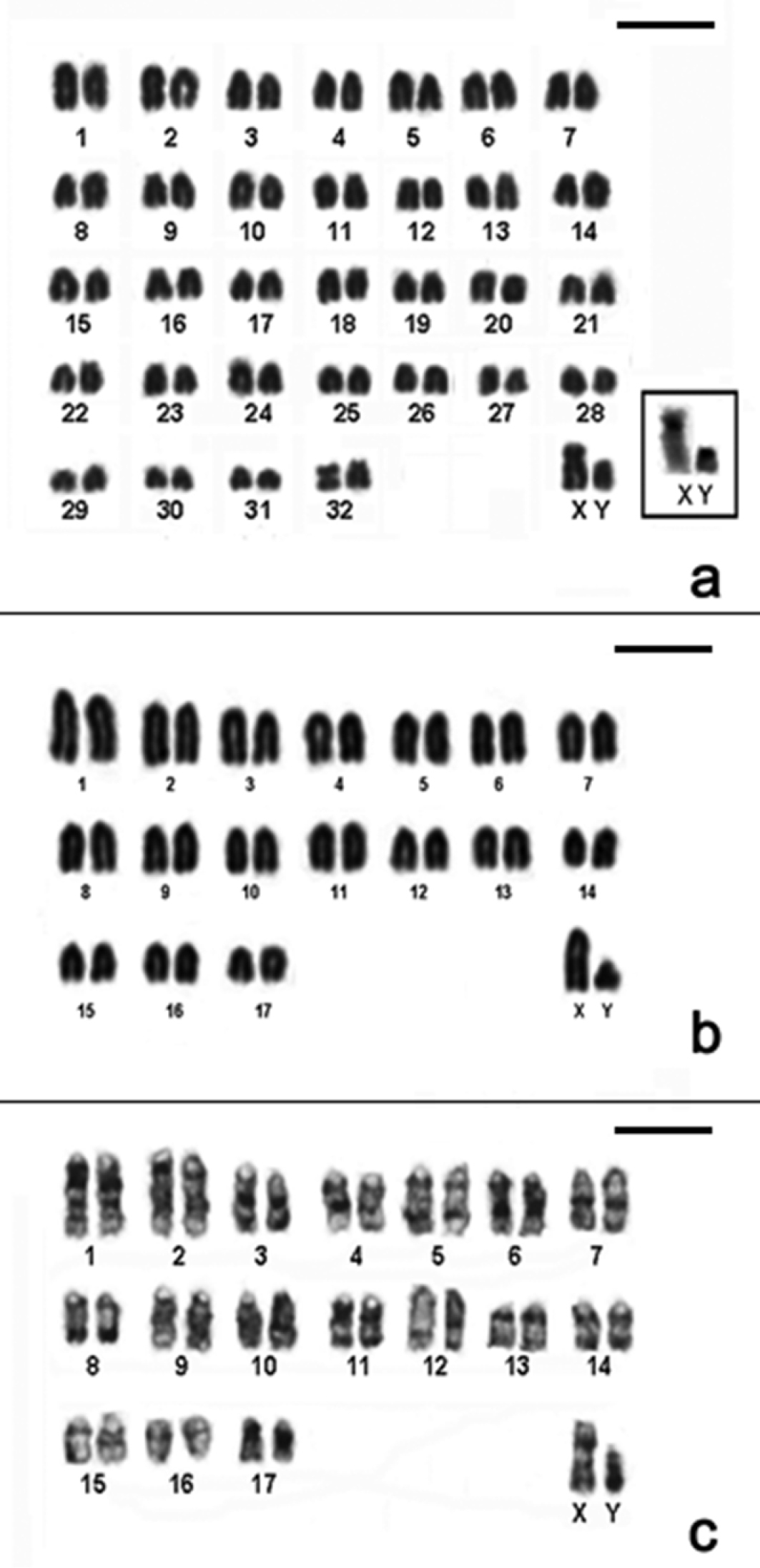
Cytogenetic data helped us to report for first time the presence of Akodon montensis, and Brucepattersonius soricinus in PESM. Cytogenetic information of these species are shown in Fig. 3, Table 1. Briefly, Akodon montensis showed 2n=24, 25 (24+1B), FNa=42 and one individual showed a heteromorphic X chromosome with an enlarged short arm. We also detected one small supernumerary submetacentric (B) in three out of nine individuals analyzed (Fig. 3a).
Karyotypes of the new records for PESM. a Conventional stained karyotype of Akodon montensis (2n=24+1B, FNa=42, male) b Conventional stained karyotype of Brucepattersonius soricinus (2n=52, FNa=52, female). Inset: sex chromosomes of a male c CBG-banding pattern of Brucepattersonius soricinus (2n=52, FNa=52, female) d GTG-banding pattern of Brucepattersonius soricinus (2n=52, FNa=52, female). Bar = 10µm.
Brucepattersonius soricinus had 2n=52, FNa=52 (Fig. 3b) and this is the first time that banding-pattern is presented in this species. The CBG-banding pattern in the female specimen showed rather pronounced amount of pericentromeric heterochromatin in all chromosomes (Fig. 3c). GTG-banding allowed the identification of all autosomic pairs and X chromosomes (Fig. 3d).
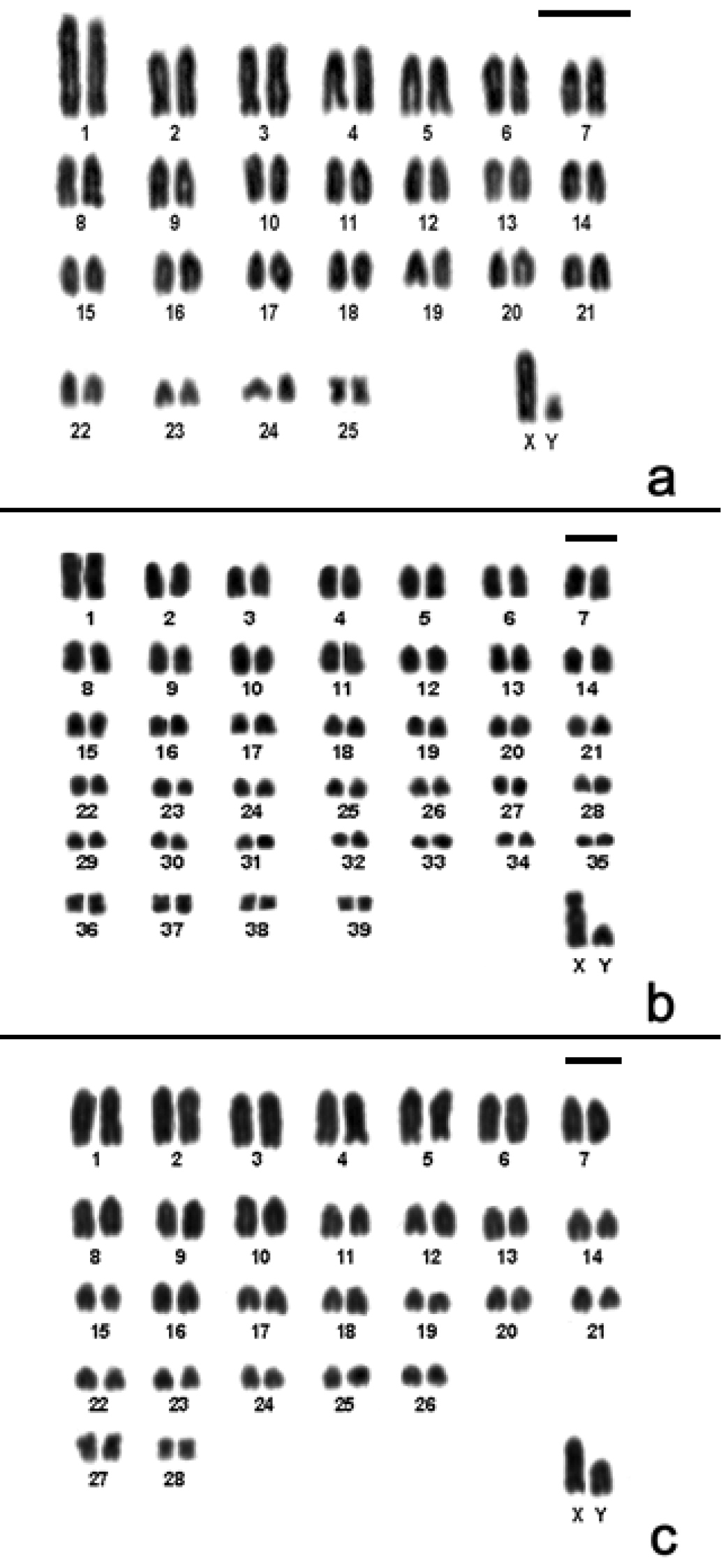
The remaining species studied in this work have already been recorded in PESM and their karyotypes are in accordance to the literature. Karyotype information of all species analyzed and the chromosomal variability found in this work is shown in Table 1 and Figs 4–7.
CBG-banding pattern of Nectomys squamipes (2n=56 + 2B, FNa=56, female) b Conventional stained karyotype of Oligoryzomys nigripes (2n=62, FNa=80, male). Inset: different forms of pair 3: heteromorphic (3H) and homomorphic metacentric (3M) c Conventional stained karyotype of Rhipidomys itoan (2n=44, FNa=50, female) d Conventional stained karyotype of Trinomys iheringi (2n=60+4Bs, FNa=116, female). Inset: sex chromosomes of a male. Bar = 10µm.
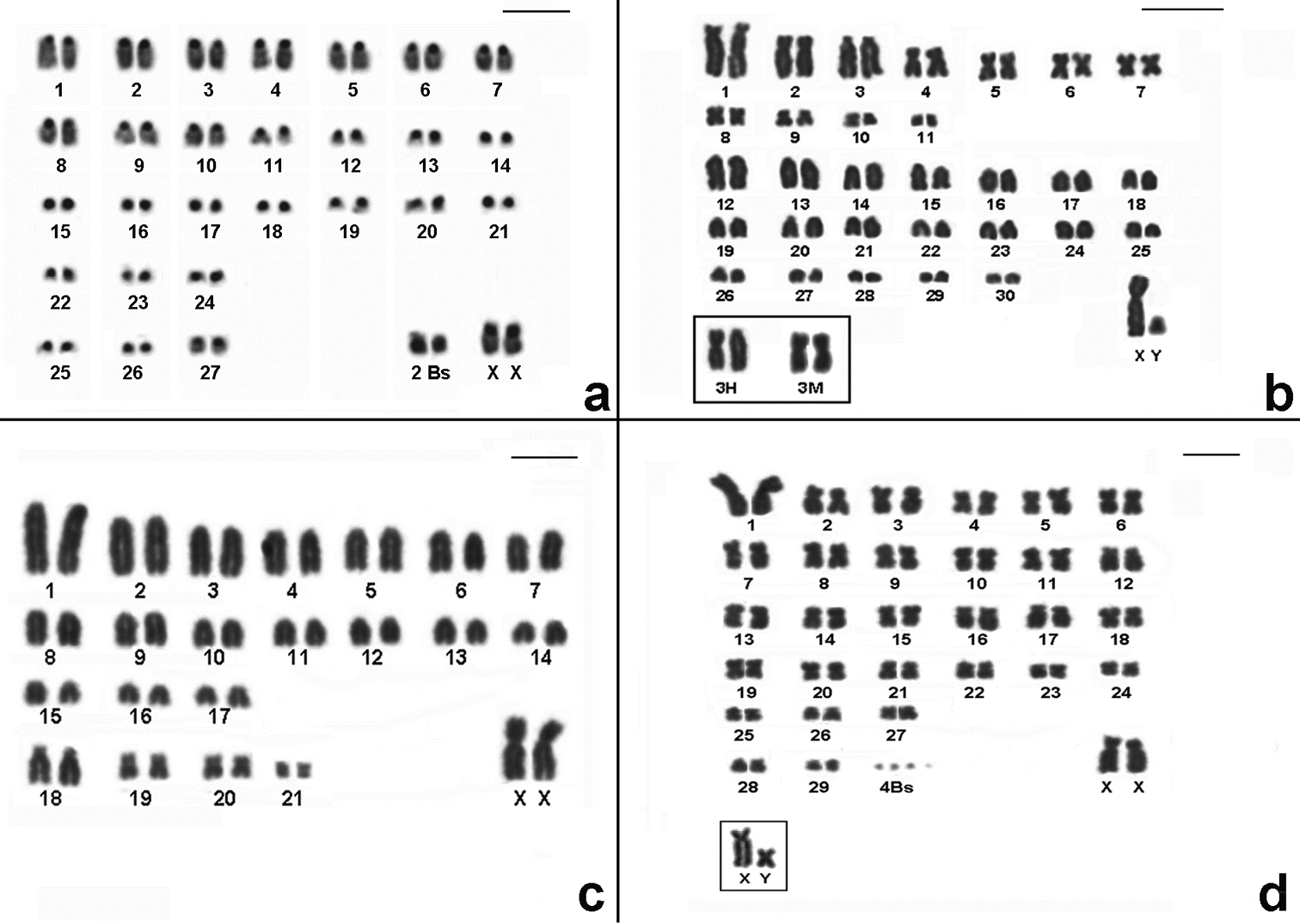
Conventional stained karyotypes: a Thaptomys nigrita (2n=52, FNa=52, male) b Euryoryzomys russatus (2n=80, FNa=86, male) c Sooretamys angouya (2n=58, FNa=60, male). Bar = 10µm.
a Conventional stained karyotype of Calomys tener (2n=66, FNa=66, male). Inset: Sex chromosomes CBG-banded b Conventional stained karyotype of Juliomys pictipes (2n=36, FNa=36, male) c GTG-banding pattern of Juliomys pictipes (2n=36, FNa=36, male). Bar = 10µm.
a CBG-banding pattern of Marmosops incanus (2n=14, FNa=24, male) b Conventional stained karyotype of Micoureus paraguayanus (2n=14, FNa=20, male) c Conventional stained karyotype of Philander frenatus (2n=22, FNa=20, male). Bar = 10µm.
Seven out of the 13 rodent species showed species-specific karyotypes: Akodon montensis, Drymoreomys albimaculatus, Oligoryzomys nigripes, Sooretamys angouya, Calomys tener, Juliomys pictipes and Trinomys iheringi (grey cells in Table 1). The identification of the remaining species (Blarinomys breviceps, Brucepattersonius soricinus, Thaptomys nigrita, Euryoryzomys russatus, Nectomys squamipes, and Rhipidomys itoan) required additional morphological and molecular investigation and geographic distribution information (Table 1).
Marsupials presented conserved diploid numbers of 14, 18 and 22 and were identified here by external morphological comparisons.
We proved the cytogenetic analyses as a taxonomic tool, since 7 out of 13 rodent species present species-specific karyotypes (53.8%). Besides, we identified 94% of all species, when cytogenetic data were combined with information of external morphology and geographical distribution (Table 1).
Cryptic species are relatively common in some Neotropical rodent groups and cytogenetic information was indispensable for identifying such species. For instance, Akodon montensis is morphologically indistinguishable from Akodon cursor (Winge, 1887) and both species occur in sympatry in the Atlantic Forest (
Another cryptic species case occurs in the genus Thaptomys. Thaptomys sp. (2n=50) and Thaptomys nigrita (2n=52) are morphologically identical, so the karyotypes are the diagnostic information to distinguish both species (
By contrast, Thaptomys nigrita and Brucepattersonius soricinus present very similar karyotypes (2n=52, FNa=52) however their identification can be safely done at the level of genera by external morphological characters. An accurate observation on the karyotypes of Brucepattersonius soricinus and Thaptomys nigrita showed that the pair 1 of Thaptomys nigrita is the largest of the chromosome set (Fig. 5a) meanwhile Brucepattersonius soricinus has the pair 1 similar in size to the others of the set (Figs 3b–d). We also noticed differences regarding sex chromosome morphologies of both species (Table 1). This feature could be a diagnostic tool to differentiate each karyotype, but additional cytogenetic studies (including comparative and molecular cytogenetic data) are needed to support these first observations.
Blarinomys breviceps presents a peculiar karyotype and it could not be considered species-specific due to the great variability in 2n and FNa (
Euryoryzomys russatus does not have species-specific karyotype also. Euryoryzomys emmonsae Musser, Carleton, Brothers and Gardner, 1998, and Euryoryzomys nitidus (Thomas, 1884) share the same 2n=80, NFa=86 (
Concerning Nectomys squamipes, it is not possible to affirm that this species possess species-specific karyotype with classical cytogenetic data because, when compared to Holochilus brasiliensis (Desmarest 1819), both karyotypes are identical (
The karyotype of Rhipidomys itoan presented here (2n=44, FNa=50 Fig. 4c) is the same one as described by
Finally, cytogenetic analysis was useful in identifying Thaptomys iheringi as two species – Thaptomys iheringi and Thaptomys dimidiatus (Günther, 1876) - occur in Atlantic Forest. Despite the regular chromosome set of Thaptomys iheringi (not considering B chromosomes) is identical to the one described for the species Thaptomys dimidiatus (2n=60, FNa=116) by
Mammals have remarkable diversity in species karyotypes, and rodents exhibit noteworthy variability of diploid chromosome number (
The chromosome variation observed here is due to the presence of supernumerary chromosomes (B chromosomes), sex chromosome heteromorphism and/or polymorphism, as well as autosomal polymorphisms. This chromosome variability does not cause a problem in characterizing the species, except in the case of Thaptomys iheringi, in which the presence of at least one B chromosome is sufficient to confirm its identity.
Structural rearrangements may explain much of the observed karyotype diversity in rodents. In this regard, Robertsonian fusions/fissions (whole-arm translocations) and pericentric inversions, have long been considered the predominant rearrangements in natural populations of rodents (
Our data showed two species with pericentric inversion rearrangements, Oligoryzomys nigripes and Rhipidomys itoan. Oligoryzomys nigripes showed variation in autosomal pair 3 (Fig. 4b) but this rearrangement had also been reported in pairs 2, 4 and 8, which places this species as one of the most polymorphic within Neotropical rodents (
Karyotype diversity is also enhanced in mammals due to the presence of B chromosomes. B chromosomes are extra elements found in the karyotypes of many eukaryotic species. Their functions and molecular composition remain obscure but, apparently in mammals, these chromosomes neither promote phenotypic alterations nor affect fitness of individuals (
Sex chromosome heteromorphisms/polymorphisms were found in Akodon montensis and Oligoryzomys nigripes, and the variation is due to addition/deletion of constitutive heterochromatin, as described by
Cytogenetic data exposed three diploid numbers for the family Didelphidae: 2n=14, 18 and 22 (
In the present paper we report for the first time the karyotype of Monodelphis scalops which is similar to the one described for Monodelphis kunsi Pine, 1975 and Monodelphis brevicauda (Erxleben, 1777) by
Our species list is an evidence of the limited knowledge of small mammals in PESM since the karyotype of Monodelphis scalops is reported for the first time and Akodon montensis and Brucepattersonius soricinus are new records for the park. According to
The improvements to the list of mammals of PESM could be attributed to different methods of capture (live and pitfall traps) to enhance the success of trapping in different habitats. The multidisciplinary approach employed is also evidently important in some cases as presented above. Additionally, data on diversity and geographical distribution of species are essential to reach conservation strategies, and the significance of Santa Virgínia / PESM in the preservation of the Neotropical fauna becomes more clear.
The authors are greatly indebted to the head of Santa Virgínia (João Paulo Villani), Dr. Mauro Galetti for supporting CLN field work, Adelita Santiago for technical support, and Elkin Suárez-Villota for critical review of the early version of the manuscript. This work was supported by the Program “Jovem Pesquisador” FAPESP for MJJS (Proc. 05/04557-3) and BIOTA-FAPESP for MG (Proc. 07/03392-6). Grants for CBDN (FAPESP Proc. 08/00493-9 and 10/03432-0), CLN (CNPq Proc. 131891/2008-1), and JFV (101.822/2011 FAPERJ) are also acknowledged. We thank the anonymous reviewers and the editor for their helpful comments.