Citation: Via Do Pico GM, Dematteis M (2014) Cytotaxonomy of two species of genus Chrysolaena H. Robinson, 1988 (Vernonieae, Asteraceae) from Northeast Paraguay. Comparative Cytogenetics 8(2): 125–137. doi: 10.3897/CompCytogen.v8i2.7209
Chromosome counts and karyotypes of two species of Chrysolaena H. Robinson 1988 are presented in this paper. Mitotic analysis revealed that both taxa have x=10, a basic chromosome number considered characteristic of the genus. The chromosome number and the karyotype of Chrysolaena cristobaliana are reported for the first time, as well as a new cytotype and the karyotype of Chrysolaena sceptrum. Chrysolaena cristobaliana showed heptaploid cytotype with 2n=7x=70 and a karyotype composed of 46 m + 24 sm chromosomes. On the other hand, Chrysolaena sceptrum presented tetraploid cytotype with 2n=4x=40 and a karyotype with 30 m + 10 sm chromosomes. Accessory chromosomes were observed in cells of both species. The chromosome analysis showed that these species differ in the chromosome number and the total chromosome length, although they showed similar chromosome morphology and asymmetry indexes. The results support the use of chromosome data in taxonomic treatments of the American members of the tribe Vernonieae.
B chromosomes, chromosome numbers, karyotype, Lepidaploinae, polyploidy
The genus Chrysolaena (Vernonieae, Asteraceae) includes 18 species mainly concentrated in southern Brazil and northeast of Argentina. From this area, the genus extends to north Peru and the Amazon region of Brazil, and southward, to the center of the province of Buenos Aires in Argentina. Most of the species of Chrysolaena occur in Brazil (15 spp.), Paraguay (11 spp.) and Argentina (6 spp.). However, a small number of species are found in Bolivia, Peru and Uruguay. Species of the genus are characterized by sericeous or velutinous indumentum, glandular anthers appendages, style without a basal node and glandular cypselas (
Since the taxonomic treatment realized by
Chrysolaena cristobaliana Dematt., 2009, and Chrysolaena sceptrum are erect shrubs with well-developed xylopodia and its distribution is mostly restricted regarding to the other species of the genus. Both taxa grow on high fields and “Cerrados” from northeast of Paraguay and southeastern Mato Grosso and Mato Grosso do Sul in Brazil (
In the present study, Chrysolaena cristobaliana and Chrysolaena sceptrum were cytologically examined in order to extend the cytogenetic knowledge and provide information taxonomically useful. The chromosome number and the karyotype of Chrysolaena cristobaliana are reported for the first time, as well as a new cytotype and the karyotype of Chrysolaena sceptrum.
The specimens were obtained from natural populations from department of Amambay, northeast of Paraguay. Voucher specimens are kept at the herbarium of the Instituto de Botánica del Nordeste (CTES). Location and Voucher specimens: Chrysolaena cristobaliana: Paraguay, Dpto. Amambay: Chirigüelo, 2 km W Pedro Juan Caballero. Cerrado degraded, near neighborhood. Dematteis and Vega 4283, (CTES). Chrysolaena sceptrum: Paraguay, Dpto. Amambay: Chirigüelo, 2 km W Pedro Juan Caballero. Cerrado degraded, near neighborhood. Dematteis and Vega, 4289 (CTES).
Mitotic chromosome preparations were made from root meristems obtained from germinating seeds. The roots were pretreated for about 5 h in 0.002 M 8-hydroxyquinoline solution at room temperature, fixed in 3:1 absolute alcohol/acetic acid, and then stained using Feulgen’s technique. Permanent microscope slides were prepared by mounting in Euparal.
At least 10 metaphases were drawn for each population using a Zeiss camera lucida (Carl Zeiss, Germany), selecting the best for measurements. The nomenclature used to describe the chromosome morphology was the one proposed by
The following karyological parameters were evaluated: total karyotype length (TKL), centromeric index (i), chromosome length (c), arm ratio (ar), and their averages (I, C and AR, respectively); in addition, the ratio between the smallest and the largest chromosome (R</>) was calculated. The karyotype asymmetry was estimated using intrachromosomal (A1) and interchromosomal (A2) indexes suggested by
The species analyzed, the somatic chromosome numbers, the ploidy level, and the karyotypic parameters calculated are detailed in Table 1.
Chromosomal number, ploidy level, karyotype formula, total karyotype length (TKL), average chromosome length (C), average centromeric index (I), average arm ratio (AR), ratio between the smallest and the largest chromosome (R</>), intrachromosomal asymmetry index (A1) and interchromosomal asymmetry index (A2), symmetry classes of Stebbins (SC) of the Chrysolaena species analyzed. SE: standard error.
| Species and voucher | Chrysolaena cristobaliana 4283 | Chrysolaena sceptrum 4289 |
|---|---|---|
| 2n | 70 | 40 |
| Ploidy | 7x | 4x |
| Karyotype formula | 2n=46m+24sm+0-7Bs | 2n=30m+10sm+0-2Bs |
| TKL ± SE (µm) | 92.42 ± 0.10 | 41.56 ± 0.09 |
| C (µm) | 2.64 | 2.08 |
| I ± SE | 39.71 ± 0.65 | 40.61 ± 0.85 |
| AR ± SE | 0.66±0.019 | 0.69±0.024 |
| R </> | 0.43 | 0.48 |
| A1 | 0.33 | 0.31 |
| A2 | 0.23 | 0.21 |
| SC | 1B | 1B |
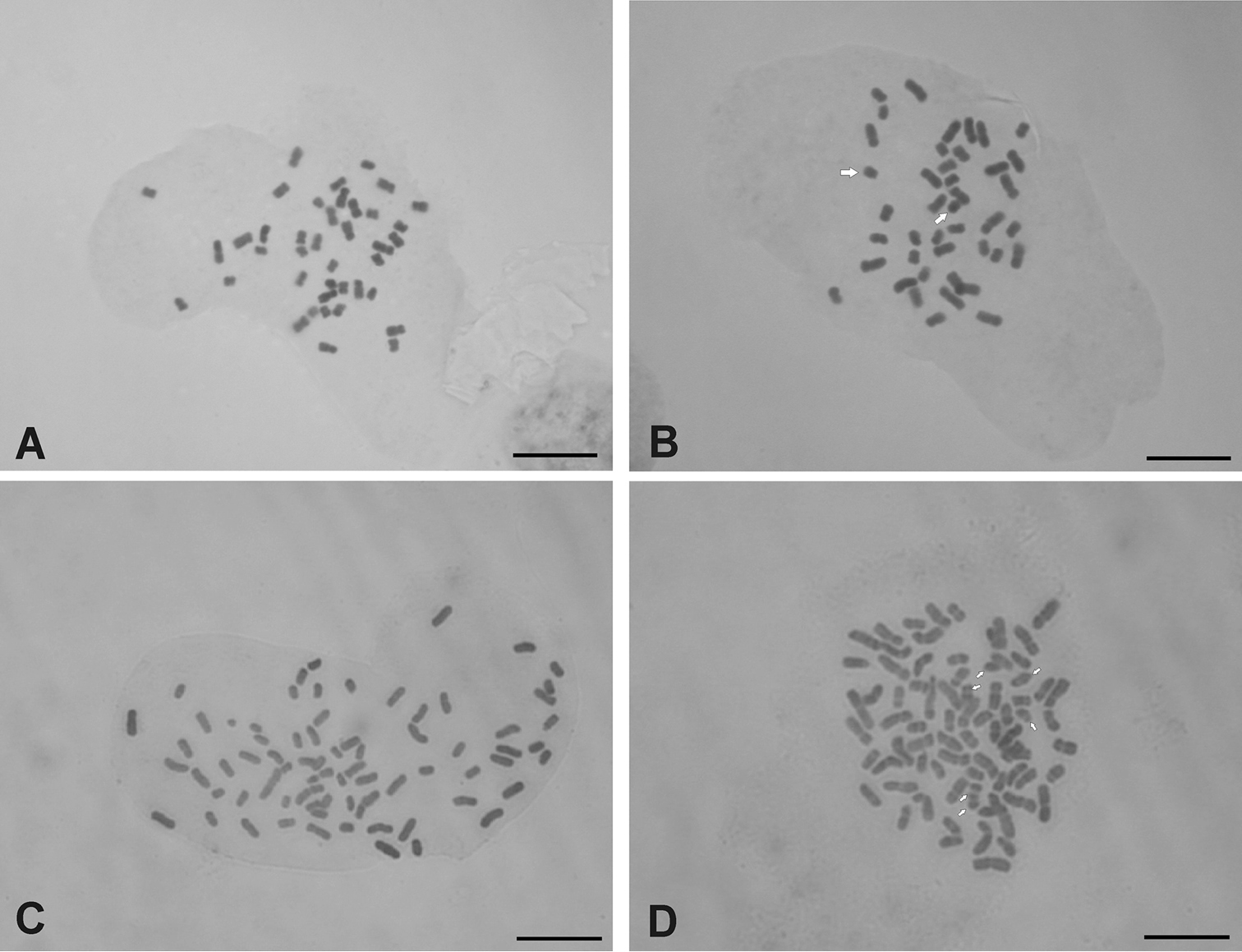
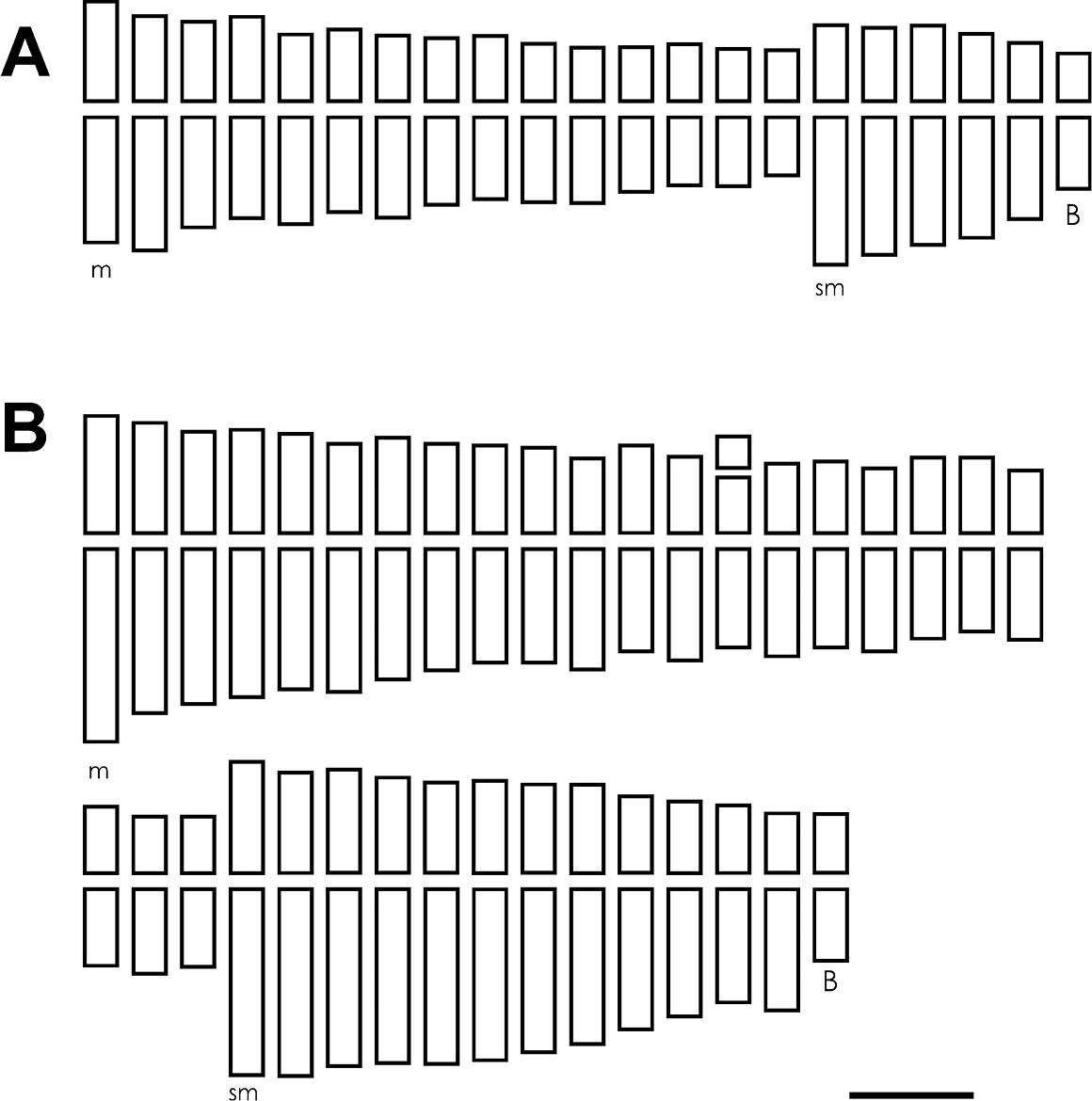
Both species analyzed presented base chromosome number x=10. Chrysolaena sceptrum showed a tetraploid cytotype with 2n=4x=40 (Fig. 1a, b), with a karyotype composed of 30 metacentric (m) and 10 submetacentric (sm) chromosomes (Figure 2a). In a few cells from 0-2 accessory or B chromosomes were observed. These elements were metacentric and showed an average size of 1.47 µm. Chrysolaena cristobaliana presented a heptaploid cytotype with 2n=7x=70, 0-7 accessory chromosomes per cell (Fig. 1c, d) and a karyotype formed by 46 (m) and 24 (sm) chromosomes (Fig. 2b). The accessory chromosomes displayed a metacentric morphology, and an average size of 1.61 µm. A single secondary constriction was observed in pair N° 25.
Figure 1. A–D Somatic chromosomes of Chrysolaena. A–B Chrysolaena sceptrum: A 2n=4x=40 B 2n=4x=40+2 Bs C–D Chrysolaena cristobaliana: C 2n=7x=70 D 2n=7x=70+6 Bs. Bar= 5 µm. White arrows denote B-chromosomes.
Figure 2. A–B Ideograms. A Chrysolaena sceptrum: 2n=30m+10sm+0-2 Bs B Chrysolaena cristobaliana: 2n=46m+24sm+0-7 Bs. Bar= 1.5 µm.
Both taxa showed moderately symmetrical karyotypes. The majority of chromosomes were metacentric, with fewer submetacentric pairs. Gradual differences in chromosome size were observed. Chrysolaena cristobaliana showed an average centromeric index I=39.71, an asymmetry index A1=0.33 and A2=0.23; while Chrysolaena sceptrum presented values of I= 40.61, A1= 0.31 and A2= 0.21. The average chromosome length (C) in Chrysolaena cristobaliana was 2.64 µm, while in Chrysolaena sceptrum it was 2.08 µm (see Table 1). According to the classification of
In this study we reported for the first time the chromosome number and the karyotype of Chrysolaena cristobaliana. Besides, the karyotype of Chrysolaena sceptrum was recorded for the first time, as well as a new cytotype.
The base chromosome number x=10 is considered characteristic of Chrysolaena and clearly distinguishes this genus from the remaining American groups of the tribe. This number also has been found in the two species here analyzed, which is consistent with previous studies carried out in others Chrysolaena species (
Chrysolaena cristobaliana has never been cytologically analyzed. This study reports as novelty the base chromosome number (x=10), the chromosome number (2n=7x=70, heptaploid) and the karyotype of the species. Chrysolaena cristobaliana and Chrysolaena sceptrum are closely related species. They are distributed in the same geographic region, and even populations of both entities can be found living in the same area. The main morphological features that differentiate these two species are the branch of the stem and the leaf shape. Chrysolaena cristobaliana presents densely branched and elliptical, lanceolate to oblanceolate leaves, whilst Chrysolaena sceptrum has single stems and narrowly lanceolate to linear leaves. The chromosome counts realized here show that these two entities can also be distinguished by the chromosome number.
Chrysolaena cristobaliana is also closely related to Chrysolaena cognata, one of the most widely distributed species of the genus. However, both species differ in the leaf shape, the pubescence type, the florets number and the geographical distribution. The results of this study added the chromosome number as a feature to distinguish these two closely related species.
In species of plants and animals, that carries B chromosomes, those individuals from a given population with and without Bs, cannot generally be phenotypically distinguished from each other. However, in some species, there are instances in which B chromosomes change certain morphological characteristics (
Chrysolaena cristobaliana and Chrysolaena sceptrum were never been karyotypically characterized. Karyotype analysis is essential for the cytogenetic characterization of species and to examine the variation between its individuals and/or populations. The comparison of karyotypes of different species also allows the taxonomic and evolutionary analysis of a taxon, such as a genus. Besides, many times, differences in karyotype asymmetry can indicate how these chromosomes have diversified in size and morphology within a group (
Chrysolaena cristobaliana presents the longest karyotype (TKL), which is correlated with its ploidy level (2n=7x=70). Although the species differed in their chromosome number and total karyotype length, they had similar chromosomal morphology and asymmetry indices. Therefore, karyotype data do not seem to be of great use for group taxonomy, since the chromosomes are small and the karyotype differences cannot be detected rapidly by the analysis of one or a few cells of each species, but only by comparing average measurements. According to
The species of Chrysolaena exhibit abundant polyploidy. The majority of species studied so far, include diploid and polyploid populations. There are some exceptions of species with a single known cytotype, such as the diploid Chrysolaena verbascifolia (Dematteis 1997,
Moreover, the chromosome number found in Chrysolaena cirstobaliana is the second report of impair ploidy level for the genus Chrysolaena. In Chrysolaena cognata a mixed population with pentaploid (2n=5x=50) and hexaploid specimens (2n=6x=60) was found in Misiones, Argentina (
There are many important characters of taxonomic weight to separate Chrysolaena from other Vernonieae groups, such as the morphology of the pollen grains and the morphological characters. However, the chromosome number is considered one of the most important features, since Chrysolaena is the only American member of the tribe with the base number x=10, which is mainly present in the Old World Vernonieae. Historically, base chromosome numbers have been widely employed for the delimitation of generic and infrageneric taxa in the Compositae (
This study was supported by greatly appreciated grants from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET).