Citation: Vittorazzi SE, Quinderé YRSD, Recco-Pimentel SM, Tomatis C, Baldo D, Lima JRF, Ferro JM, Lima LD, Lourenço LB (2014) Comparative cytogenetics of Physalaemus albifrons and Physalaemus cuvieri species groups (Anura, Leptodactylidae). Comparative Cytogenetics 8(2): 103–123. doi: 10.3897/CompCytogen.v8i2.6414
Recently, Physalaemus albifrons (Spix, 1824) was relocated from the Physalaemus cuvieri group to the same group as Physalaemus biligonigerus (Cope, 1861), Physalaemus marmoratus (Reinhardt & Lütken, 1862) and Physalaemus santafecinus Barrio, 1965. To contribute to the analysis of this proposition, we studied the karyotypes of Physalaemus albifrons, Physalaemus santafecinus and three species of the Physalaemus cuvieri group. The karyotype of Physalaemus santafecinus was found to be very similar to those of Physalaemus biligonigerus and Physalaemus marmoratus, which were previously described. A remarkable characteristic that these three species share is a conspicuous C-band that extends from the pericentromeric region almost to the telomere in the short arm of chromosome 3. This characteristic is not present in the Physalaemus albifrons karyotype and could be a synapomorphy of Physalaemus biligonigerus, Physalaemus marmoratus and Physalaemus santafecinus. The karyotype of Physalaemus santafecinus is also similar to those of Physalaemus marmoratus and Physalaemus biligonigerus owing to the presence of several terminal C-bands and the distal localization of the NOR in a small metacentric chromosome. In contrast, the Physalaemus albifrons karyotype has no terminal C-bands and its NOR is located interstitially in the long arm of submetacentric chromosome 8. The NOR-bearing chromosome of Physalaemus albifrons very closely resembles those found in Physalaemus albonotatus (Steindachner, 1864), Physalaemus cuqui Lobo, 1993 and some populations of Physalaemus cuvieri Fitzinger, 1826. Additionally, the Physalaemus albifrons karyotype has an interstitial C-band in chromosome 5 that has been exclusively observed in species of the Physalaemus cuvieri group. Therefore, we were not able to identify any chromosomal feature that supports the reallocation of Physalaemus albifrons.
Chromosome, NOR, C-banding, heterochromatin, Physalaemus
Currently, the genus Physalaemus Fitzinger, 1826 is classified in the subfamily Leiuperinae Bonaparte, 1850 in the family Leptodatylidae Werner, 1896 (
According to the taxonomic proposal of
Detailed descriptions of the karyotypes of Physalaemus biligonigerus and Physalaemus marmoratus (as Physalaemus fuscomaculatus), which included the identification of the nucleolus organizer regions (NOR) and heterochromatic sites, were already provided (
In the present work, we present a detailed characterization of the karyotype of Physalaemus albifrons, describe the karyotype of Physalaemus santafecinus and extend the cytogenetic analyses of the Physalaemus cuvieri group in order to better characterize the group from which Physalaemus albifrons was removed. Our aim is to provide additional evidence that could be used to compare the Physalaemus albifrons and Physalaemus cuvieri species groups.
Specimens of Physalaemus albifrons, Physalaemus santafecinus, Physalaemus albonotatus (Steindachner, 1864), Physalaemus centralis Bokermann, 1962 and Physalaemus cuqui Lobo, 1993 from different localities in Brazil and Argentina were analyzed. For an unequivocal identification of the species, both morphological and acoustic characteristics were utilized. Each specimen’s locality and voucher number in the scientific collection where it was deposited are provided in Table 1.
Locality, voucher number and chromosome location of NORs and C-bands of the analyzed specimens. Abbreviations: CH – Chaco province; CT – Corrientes province; MA – Maranhão state; MT - Mato Grosso state; SA – Salta province; SP - São Paulo state; TO - Tocantins state; BC - Departamento de Biologia Celular da UNICAMP, Campinas, SP, Brazil; LGE - Laboratorio de Genética Evolutiva, Facultad de Ciencias Exactas Químicas y Naturales, Universidad Nacional de Misiones, Posadas, Misiones, Argentina; MNRJ - Museu Nacional do Rio de Janeiro, RJ, Brazil; UFMT - Universidade Federal do Mato Grosso, MT, Brazil; ZUEC - Museu de História Natural, Universidade Estadual de Campinas, Campinas, SP, Brazil; p: short arm; q: long arm; int: interstitial; per: pericentromeric; 3cen-per: centromeric band that extend to the short arm. * only one chromosome of the pair. **In the ZUEC 13696 specimen, an additional terminal C-band is present in 2q (see text for details).
| Species | Locality | Specimens | NOR locations | Principal non-centromeric C-bands |
|---|---|---|---|---|
| Physalaemus albifrons | Barreirinhas, MA, Brazil | 7 ♂ (MNRJ 24228, 24230, 24232, ZUEC 12361–3, 17925), 1♀ (MNRJ 24227) | 8q | 3cen-per/5p int/8p per/ 9p per |
| Physalaemus albonotatus | Lambari D´Oeste, MT, Brazil (57.4°W, 16.4°S) | 6 ♂ (UFMT 4462, 4466, 4469–72), 1♀ (UFMT 4465) | 8q/9p/9q | 2q int/3cen-per/5 int/ |
| Physalaemus centralis | Palestina, SP, Brazil (49.2°W, 20.2°S) | 5 ♂ (ZUEC 13689–90, 13692, 3694, 13696) | 9q per | 2q int**/3cen-per/5p int/ 8q int/9q int/10p per |
| Porto Nacional, TO, Brazil (48.6°W, 10.4°S) | 3 ♂ (ZUEC 13373, 13375, 13380) | 9q per | 2q int/3cen-per/5p int/ 8q int/9q int/10p per | |
| Physalaemus cuqui | Near to Rio Piedras, Iruya, SA, Argentina (22°56'S, 64°39'W) | 1♀ (LGE 6567) | 3p*/8q/9p/9q | 2q int/3cen-per/5p int |
| Taco Pozo, CH, Argentina (25°34'S, 63°09'W) | 2 ♂ (LGE 1635–6) | 8q/9p/9q | 2q int/3cen-per/5p int | |
| Aguas Blancas, SA, Argentina (22°43'S, 64°22'W) | 1♀ (LGE 6568) | 8q/9p/9q | - | |
| Metán, SA, Argentina (25°06'S, 65°03'W) | 1♀ (LGE 6569) | 8q*/9p/9q | 2q int/3cen-per/5p int | |
| Pichanal, SA, Argentina (25°24'S, 64°09'W) | 1♂ (LGE 6570) | 8q/9p/9q | 2q int/3cen-per/5p int | |
| Physalaemus santafecinus | Ituzaingó, CT, Argentina (27°31'S, 56°40'W | 6 ♂ (LGE 077–8, 083–4, 087–8) | 9q | 1p per/1q int/2p per/3p/8p per/7q per/terminal in all chromosomes |
Metaphase chromosome spreads were obtained from cell suspensions of the intestine and testes of animals pre-treated with colchicine (2%) for at least 4 hours (according to
All of the analyzed individuals had a diploid complement of 22 chromosomes. By comparing all of the karyotypes of Physalaemus to each other, we noted a high interspecific similarity for the first seven chromosome pairs, and the homeology of these chromosomes could be inferred. Therefore, in each karyotype presented here, these chromosomes were ordered in such a way that their numbers could reflect these homology hypotheses even when their sizes suggested a different numbering. However, the smallest chromosomes (pairs 8–11) varied significantly among the species analyzed, and were numbered only by chromosome size. A detailed description of the karyotype of each species is presented below and the Appendix (Additional file 1) present all the karyotypes arranged together. Table 1 summarizes the data on NORs and non-centromeric C-bands.
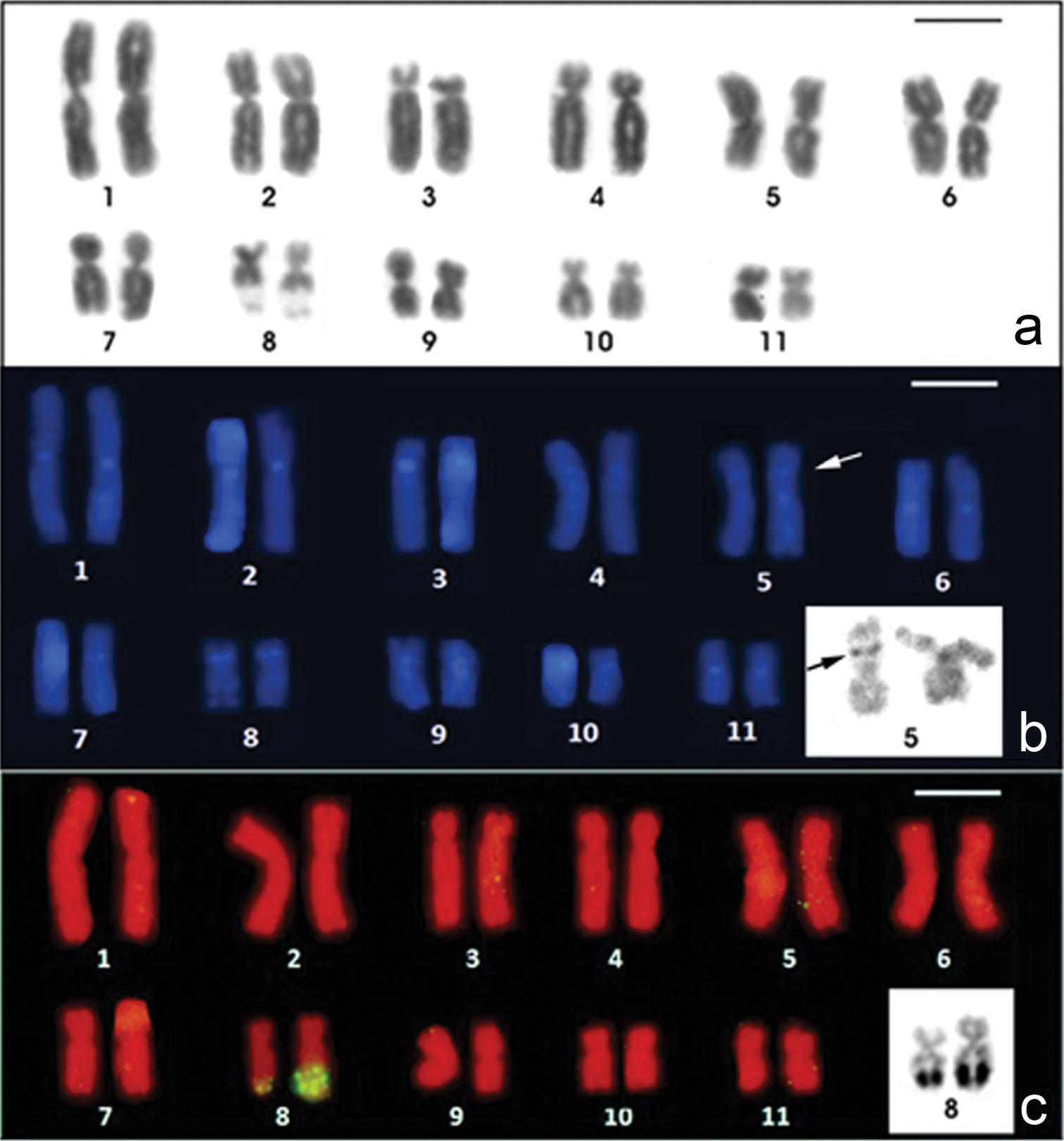
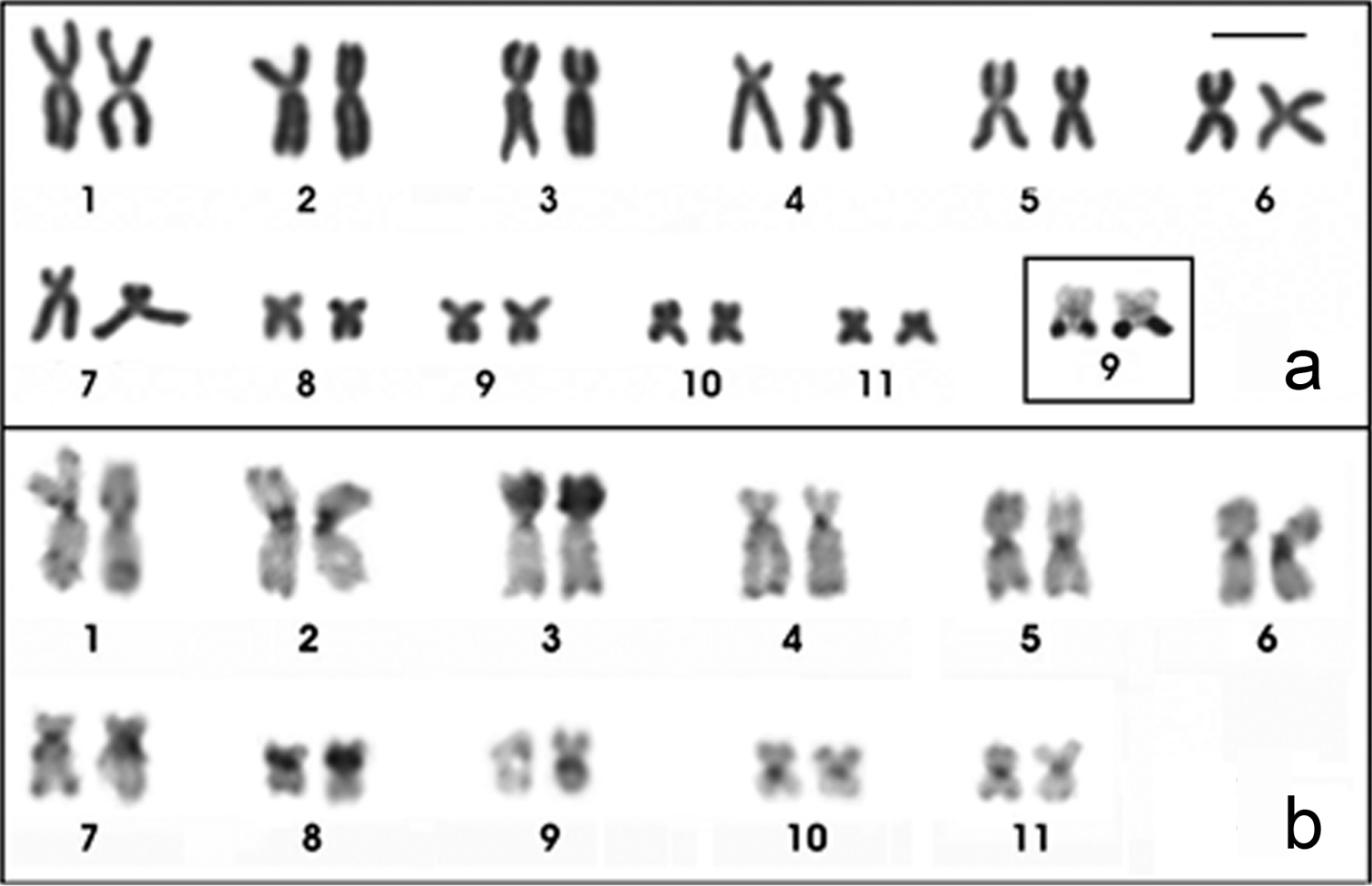
The Physalaemus albifrons chromosomes were classified as metacentric (pairs 1, 2, 5, 6, 9 and 11), submetacentric (pairs 4, 7, 8 and 10) or subtelocentric (pair 3, which is at the threshold between submetacentric and subtelocentric classifications) (Fig. 1a; Table 2). C-banding followed by DAPI staining detected all of the centromeric regions and an interstitial heterochromatic band in the short arm of chromosome 5 as well as pericentromeric bands in the short arm of chromosomes 8 and 9 (Fig. 1b). The Giemsa stained C-banded metaphases showed this same pattern, but after DAPI staining, the bands could be more easily visualized. Chromosomes 3 and 4 were very similar, but chromosome 3 had a slightly smaller centromeric index and a strong centromeric C-band, which extended to the short arm (Fig. 1b; Table 2).
Karyotypes of Physalaemus albifrons after Giemsa-staining (a) C-banding followed by DAPI-staining (b) and in situ hybridization with a nucleolar rDNA probe (c). In b the inset shows the C-banded chromosome pair 5 stained with Giemsa; in c the inset shows the NOR-bearing chromosome pair 8 after silver staining. Arrows in b point the interstitial C-band in 5p. Bar=10mm.
Morphometric parameters of the Physalaemus albifrons, Physalaemus albonotatus, Physalaemus centralis, Physalaemus cuqui and Physalaemus santafecinus karyotypes. The measurements were based on 10 metaphases of each species. CN: chromosome number; CI: centromeric index; SD: standard deviation; RL: relative length. CC: chromosome classification; m: metacentric; sm: submetacentric; st: subtelocentric. *1Chromosomes were numbered in order to reflect our hypotheses of homeology for the Physalaemus chromosomes even when their sizes suggested a different numbering. *2Value at the threshold between submetacentric and subtelocentric classifications.
| Physalaemus albifrons | |||||||||||
| CN | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| CI ± SD | 0.47 ± 0.03 | 0.40 ± 0.04 | 0.24 ± 0.02 | 0.29 ± 0.02 | 0.46 ± 0.02 | 0.44 ± 0.04 | 0.36 ± 0.02 | 0.33 ± 0.04 | 0.43 ± 0.04 | 0.28 ± 0.04 | 0.45 ± 0.04 |
| RL(%) | 14.68 | 12.15 | 10.06*1 | 10.64*1 | 9.68 | 9.43 | 8.27 | 7.17 | 6.76 | 5.94 | 5.88 |
| CC | m | m | st* | sm | m | m | sm | sm | m | sm | m |
| Physalaemus albonotatus | |||||||||||
| CN | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| CI ± SD | 0.46 ± 0.03 | 0.45 ± 0.04 | 0.24 ± 0.02 | 0.33 ± 0.02 | 0.46 ± 0.03 | 0.43 ± 0.04 | 0.36 ± 0.07 | 0.39 ± 0.04 | 0.44 ± 0.03 | 0.42 ± 0.03 | 0.46 ± 0.03 |
| RL(%) | 13.87 | 12.18 | 10.00*1 | 10.42*1 | 9.61 | 9.48 | 8.31 | 7.32 | 7.05 | 5.98 | 5.78 |
| CC | m | m | st* | sm | m | m | sm | m | m | m | m |
| Physalaemus centralis | |||||||||||
| CN | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| CI ± SD | 0.47 ± 0.01 | 0.39 ± 0.01 | 0.26 ± 0.02 | 0.30 ± 0.03 | 0.46 ± 0.03 | 0.43 ± 0.03 | 0.35 ± 0.03 | 0.42 ± 0.05 | 0.45 ± 0.01 | 0.40 ± 0.02 | 0.40 ± 0.04 |
| RL(%) | 13.82 | 12.24 | 10.07*1 | 10.26*1 | 10.03 | 9.36 | 7.99 | 7.27 | 7.12 | 6.31 | 5.52 |
| CC | m | m | sm | sm | m | m | sm | m | m | m | m |
| Physalaemus cuqui | |||||||||||
| CN | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| CI ± SD | 0.47 ± 0.02 | 0.41 ± 0.03 | 0.24 ± 0.05 | 0.30 ± 0.03 | 0.44 ± 0.03 | 0.41 ± 0.02 | 0.34 ± 0.03 | 0.42 ± 0.01 | 0.42 ± 0.06 | 0.38 ± 0.01 | 0.43 ± 0.03 |
| RL(%) | 14.53 | 13.57 | 10.0*1 | 10.36*1 | 9.93 | 9.49 | 8.39 | 7.08 | 6.07 | 5.40 | 5.19 |
| CC | m | m | st* | sm | m | m | sm | m | m | m | m |
| Physalaemus santafecinus | |||||||||||
| CN | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| CI ± SD | 0.46 ± 0.02 | 0.40 ± 0.01 | 0.39 ± 0.02 | 0.27 ± 0.02 | 0.46 ± 0.02 | 0.43 ± 0.01 | 0.32 ± 0.02 | 0.39 ± 0.03 | 0.47 ± 0.01 | 0.43 ± 0.03 | 0.43 ± 0.04 |
| RL(%) | 14.11 | 13.21 | 12.34 | 10.88 | 10.27 | 9.67 | 8.60 | 5.68 | 5.41 | 5.35 | 4.47 |
| CC | m | m | m | sm | m | m | sm | m | m | m | m |
The NORs were located distally in the long arm of chromosome 8 (Fig. 1c) and coincided with the secondary constrictions that were observed in Giemsa-stained metaphases (Fig. 1a). In three specimens (ZUEC 17925, ZUEC 12363 and MNRJ 24224), a size heteromorphism was observed between the homologous NORs by FISH with an rDNA probe (Fig. 1c) and by silver staining (Fig. 1c - inset). In two specimens (MNRJ 24230 and 24232), the NOR-bearing homologous chromosomes 8 were homomorphic. For the remaining specimens, we were not able to determine if a NOR size heteromorphism was present.
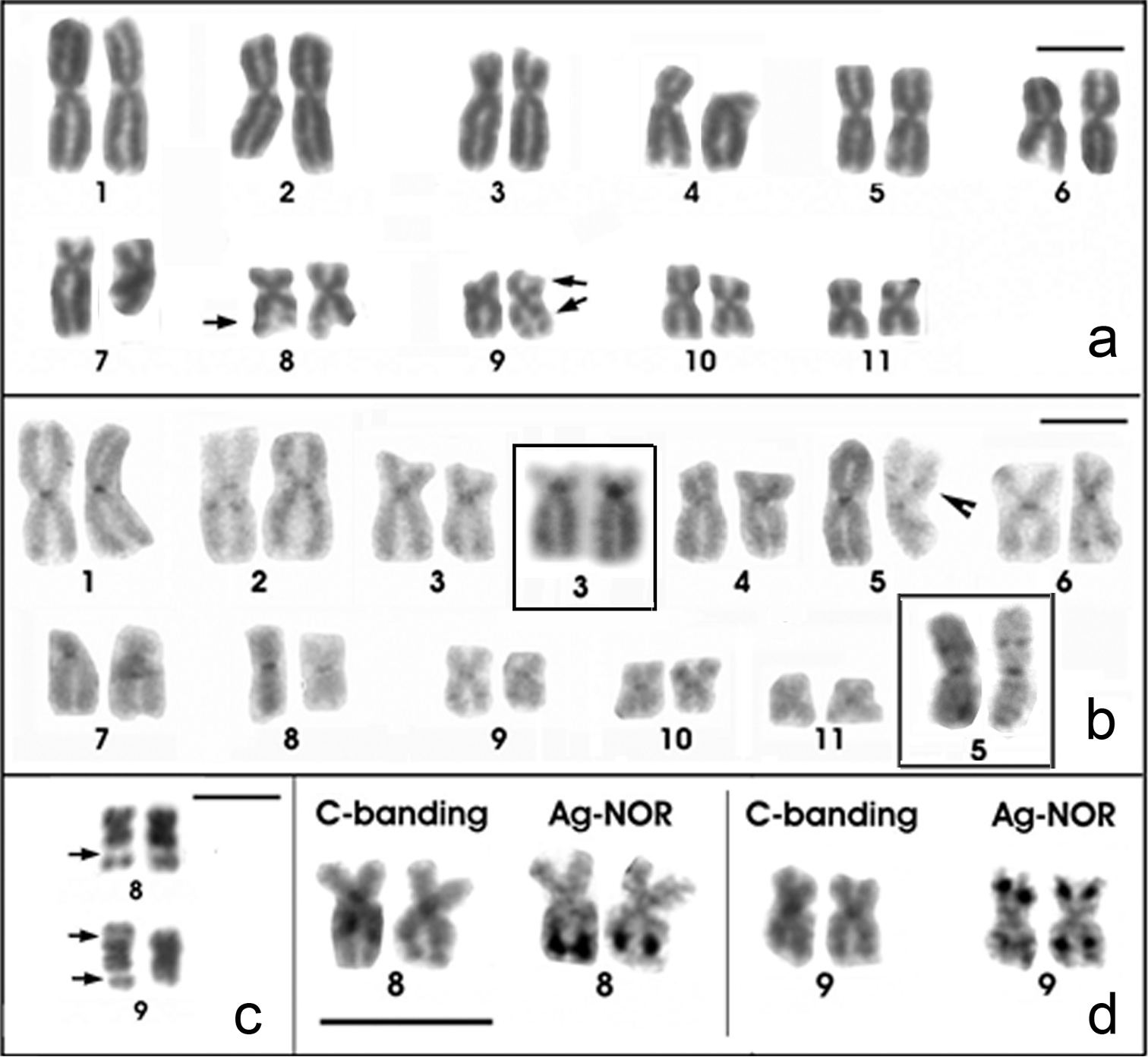
The Physalaemus albonotatus chromosomes were classified as metacentric (pairs 1, 2, 5, 6, 8, 9, 10 and 11), submetacentric (pairs 4 and 7) or subtelocentric (pair 3, which is at the threshold between submetacentric and subtelocentric classifications) (Fig. 2a; Table 2). Curiously, chromosome 5 was larger than chromosomes 3 and 4 in some of the analyzed metaphases (as seen in Figure 2b). Heterochromatin was detected in the centromeres of all chromosomes and interstitially in the long arm of chromosome 2 and in the metacentric chromosome 5 (Fig. 2b). Only two C-banded chromosome pairs 5 were good enough to be measured. Therefore, we tentatively assigned the interstitial C-band of chromosome 5 to its short arm, but further analyses are necessary to test this hypothesis. Chromosomes 3 and 4 were very similar, but chromosome 3 had a slightly smaller centromeric index and a strong centromeric C-band, which extended to the short arm (Fig. 2b - inset; Table 2).
Giemsa-stained (a) and C-banded (b) karyotypes of Physalaemus albonotatus. In the insets in b C-banded chromosome pairs 3 and 5, showing evident pericentromeric and interstitial bands, respectively c NOR-bearing chromosome pairs of Physalaemus albonotatus stained with Giemsa. Arrows in a and c indicate secondary constrictions of the NORs. Arrowhead in b indicates the C-band in chromosome 5 d NOR-bearing chromosome pairs of one specimen of Physalaemus albonotatus sequentially submitted to the C-banding and the Ag-NOR methods. Note the NOR adjacent to an interstitial C-band in pair 8 and the NORs coincident with faint C-bands in pair 9. Bar=10mm.
Silver staining detected NORs distally in the long arm of chromosome 8 adjacent to a faint C-band and in both arms of chromosome 9 (Fig. 2d). The NOR in the long arm of chromosome 9 apparently coincided with a C-band (Fig. 2d). All of these NORs could be seen as secondary constrictions in Giemsa-stained metaphases (Figs. 2a and 2c).
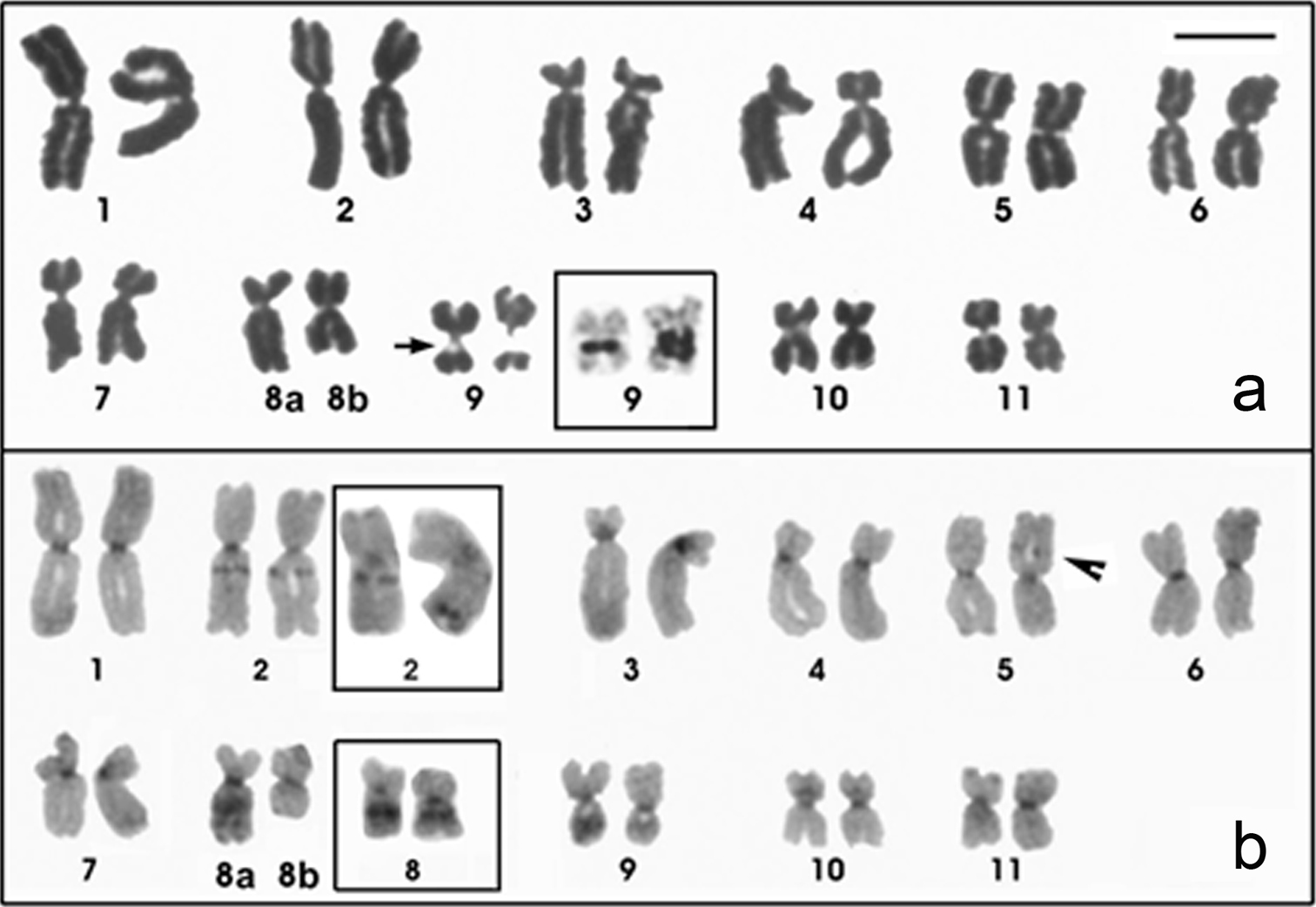
The Physalaemus centralis chromosomes were classified as metacentric (pairs 1, 2, 5, 6, 8, 9, 10 and 11) or submetacentric (pairs 3, 4, 7 and 8) (Fig. 3a; Table 2). A secondary constriction was detected in the pericentromeric region of the long arm of chromosome 9 and coincided with the NOR that was recognized by silver staining (Fig. 3a - inset). A NOR size heteromorphism was observed in all of the Physalaemus centralis specimens analyzed. C-bands were present interstitially in the long arm of chromosome 2, in the short arm of chromosome 5, in the long arms of chromosomes 8 and 9, in the pericentromeric region of the short arm of chromosome 10, and in all of the centromeres (Fig. 3b). Chromosomes 3 and 4 were very similar, but chromosome 3 had a slightly smaller centromeric index and a strong centromeric C-band, which extended to the short arm (Fig. 3b; Table 2).
Giemsa-stained (a) and C-banded (b) karyotypes of Physalaemus centralis. In a an arrow indicates the secondary constriction of the NOR and the inset shows the NOR-bearing chromosome 9 after silver staining. Note the NOR size heteromorphism. In b the arrowhead indicates the C-band in 5p and the insets show the heteromorphic pair 2 and the homomorphic pair 8 of the ZUEC 13696 specimen. Note the conspicuous interstitial heterochromatin in the long arm of chromosome pair 8. Bar=10mm.
In three specimens, a heteromorphic chromosome pair 8 composed of homologues with different morphologies and C-banding patterns was observed (Figs. 3a and 3b). While one chromosome 8 showed a conspicuous interstitial C-band that sometimes could be seen as two heterochromatic blocks (chromosome 8a in Fig. 3b), its homologue had no observable interstitial heterochromatic block (Fig. 3b). In the ZUEC 13696 specimen, the pericentromeric C-bands in the long arms of the homologous chromosomes 2 were heteromorphic in size. Additionally, the homologue that had the smaller pericentromeric C-band also had an additional and conspicuous terminal C-band in the long arm (Fig. 3b - inset).
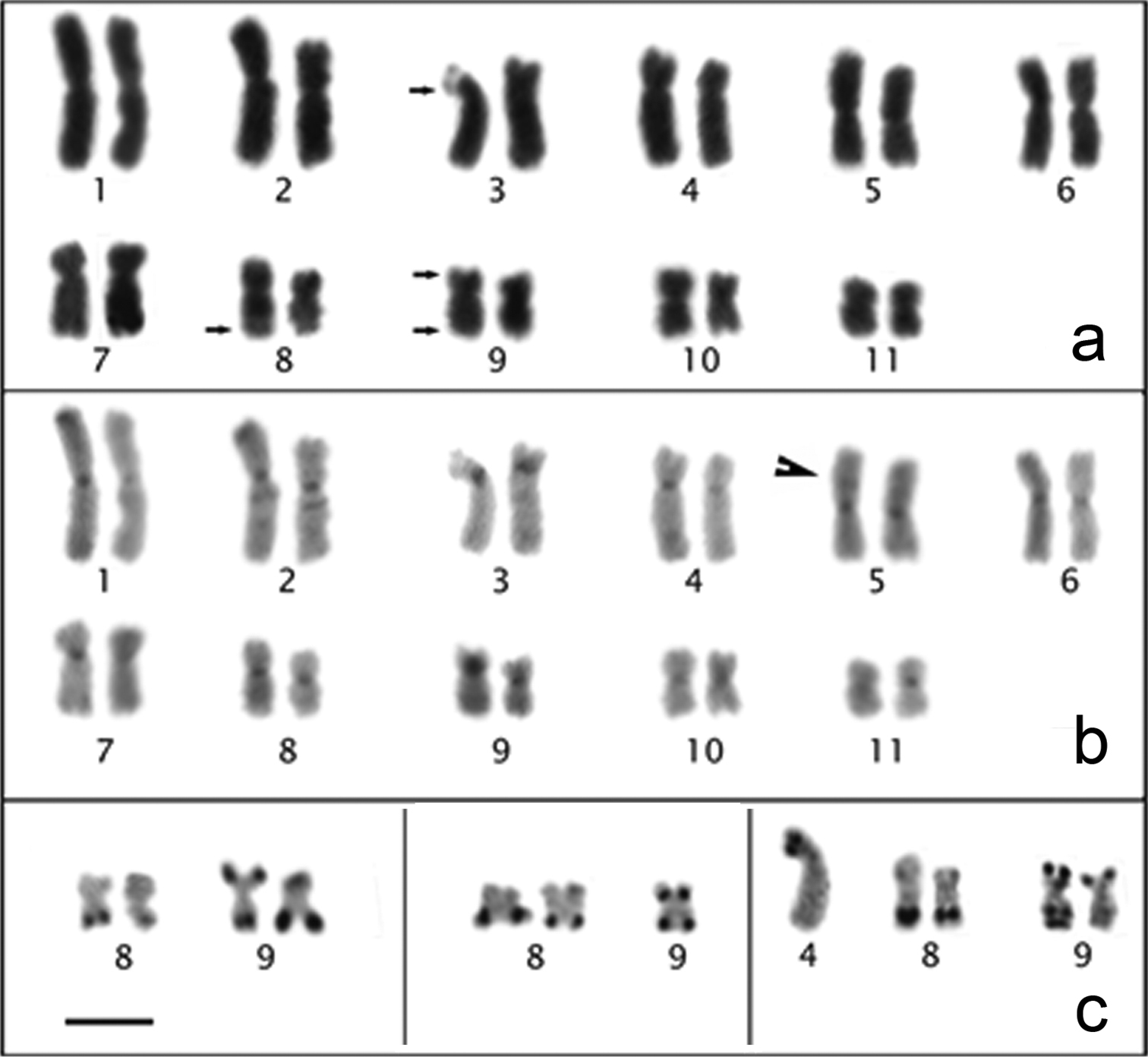
The Physalaemus cuqui chromosomes were classified as metacentric (pairs 1, 2, 5, 6, 8, 9, 10 and 11), submetacentric (pairs 4 and 7) or subtelocentric (pair 3, which is at the threshold between submetacentric and subtelocentric classifications) (Fig. 4a; Table 2). Heterochromatic bands were observed interstitially in the long arm of chromosome pair 2, in the metacentric chromosome pair 5 and in the centromeric regions of all of the chromosomes (Fig. 4b). Only one C-banded chromosome pair 5 could be measured. Therefore, as well as for Physalaemus albonotatus, we tentatively assigned the interstitial C-band of chromosome 5 of Physalaemus cuqui to its short arm, but further analyses are necessary to test this hypothesis. Chromosomes 3 and 4 were very similar, but chromosome 3 had a slightly smaller centromeric index and a strong centromeric C-band, which extended to the short arm (Fig. 4b; Table 2).
Giemsa-stained (a) and C-banded (b) karyotypes of Physalaemus cuqui. In a the arrows indicate the secondary constriction of the NOR. In b arrowhead indicates the C-band in 5p c Variability in the NOR-bearing chromosome pairs of three specimens of Physalaemus cuqui. Bar=10mm.
In three specimens, the Ag-NORs were located in the long arm of chromosome pair 8 and in the short and long arms of chromosome pair 9 (LGE 1635-1636, MLP DB 4973) (Fig. 4c – left), but only one chromosome 9 was silver-stained in the MLP DB 5560 specimen (Fig. 4c – middle). Additionally, one specimen (MLP DB 6480) showed an additional Ag-NOR in the short arm of one chromosome 4 (Fig. 4c – right). These Ag-NORs were coincident with the secondary constrictions visualized in Giemsa-stained metaphases (Fig. 4a).
The Physalaemus santafecinus chromosomes were classified as metacentric (pairs 1, 2, 3, 5, 6, 8, 9, 10 and 11) or submetacentric (pairs 4 and 7) (Fig. 5a; Table 2). The NORs were located distally in the long arm of chromosome 9 (Fig. 5a - inset). C-bands were detected in all the centromeric regions. Additionally, pericentromeric C-bands were present in the short arms of chromosomes 1 and 2 and in the short arm of chromosome 8. Small C-bands were also detected proximally in the long arms of chromosomes 4 and 7 and distally in the long arm of chromosome 1. A conspicuous C-band was observed in the short arm of chromosome 3, which was almost entirely heterochromatic. Terminal faint C-bands could be seen in all of the chromosomes (Fig. 5b). When the Ag-NOR method was performed on C-banded metaphases, we could undoubtedly recognize the chromosome 9 as the NOR-bearing chromosome while chromosomes 8 had strong pericentromeric C-bands (data not shown).
Giemsa-stained (a) and C-banded (b) karyotypes of Physalaemus santafecinus. In a, the inset shows the NOR-bearing chromosome 9 after silver staining. Bar=10mm.
To date, 23 of the 46 species of Physalaemus were karyotyped and all of them have 2n=22 (
The karyotype of Physalaemus santafecinus described here is very similar in chromosomal size and morphology to those of Physalaemus biligonigerus, Physalaemus marmoratus and Physalaemus sp. aff. biligonigerus (
A remarkable characteristic of the Physalaemus santafecinus karyotype that is shared with the karyotypes of Physalaemus biligonigerus, Physalaemus marmoratus and Physalaemus sp. aff. biligonigerus is a conspicuous C-block on the short arm of chromosome 3 (3p) (Table 3). This large heterochromatic C-block is not detected in Physalaemus albifrons or in any species of Physalaemus cuvieri group. Instead, a small C-band pericentromerically located on 3p was already detected in the karyotypes of the species currently allocated to the Physalaemus cuvieri group that were already studied by C-banding [i.e., Physalaemus albifrons, Physalaemus albonotatus, Physalaemus centralis, Physalaemus cuqui (present work), Physalaemus ephippifer (Steindachner, 1864) (
Comparison of chromosome 3 of species of Physalaemus cuvieri (left column) and P. albifrons (right column) groups. Black areas in the ideograms represent C-bands. *1Based on
Interestingly, a large 3p showing a large C-band was also observed in Physalaemus nattereri (
On the other hand, the available data do not prevent the large C-band found on 3p of Physalaemus santafecinus, Physalaemus biligonigerus and Physalaemus marmoratus from being a synapomorphy of this group of species, which could have arisen from the amplification of a small C-band. Despite the proposals of
In addition to the large C-band in 3p, the karyotype of Physalaemus santafecinus is also similar to those of Physalaemus biligonigerus, Physalaemus marmoratus and Physalaemus sp. aff. biligonigerus (
In contrast to Physalaemus santafecinus, Physalaemus biligonigerus, Physalaemus marmoratus and Physalaemus sp. aff. biligonigerus, the telomeric C-bands could not be detected in the karyotype of Physalaemus albifrons. Additionally, the NOR in Physalaemus albifrons was detected interstitially in the long arm of the submetacentric chromosome 8. This NOR-bearing chromosome very closely resembles the NOR-bearing chromosome found in some populations of Physalaemus cuvieri (
Despite the similarity between the NOR-bearing chromosome of Physalaemus albifrons and those of some species of the Physalaemus cuvieri group, it would be premature to consider this a synapomorphy of Physalaemus albifrons and species of the Physalaemus cuvieri group because the evolutionary divergence of this character (i.e., NOR location) has not yet been elucidated. We cannot discard the possibility that the NOR found in Physalaemus albifrons and in some Physalaemus cuvieri species is plesiomorphic with respect to the other NOR sites found in Physalaemus species. This interpretation derives from the fact that the NOR-bearing chromosome 8 found in other leiuperines, as Pleurodema diplolister (Peters, 1870) (Lourenço et al., 2006), resembles that of Physalaemus albifrons and some Physalaemus cuvieri species group and could constitute the same state of character.
Another chromosome feature found in Physalaemus albifrons that was also detected in species of the Physalaemus cuvieri group was the interstitial C-band in chromosome 5 (Table 4). This C-band was observed in all of the species of the Physalaemus cuvieri group already analyzed by the C-banding technique, including Physalaemus cuvieri (
Comparison of chromosome 5 of species of Physalaemus cuvieri (left column) and Physalaemus albifrons (right column) groups. Black areas in the ideograms represent C-bands. *1C-band was tentatively assigned to the short arm (see text for details). *2Based on
In conclusion, we were not able to recognize any chromosomal character that would support the reallocation of Physalaemus albifrons from the Physalaemus cuvieri group to the Physalaemus albifrons group together with Physalaemus biligonigerus, Physalaemus marmoratus and Physalaemus santafecinus.
Interestingly, in addition to the data regarding chromosomal characteristics, larval morphology also does not seem to support the composition of the Physalaemus albifrons group. Physalaemus biligonigerus, Physalaemus santafecinus and Physalaemus marmoratus have a similar larval oral disc configuration (LTRF 2/2, with a dorsal gap in the marginal papillae) that differs considerably from that of Physalaemus albifrons, whose oral disc is almost identical to that of the tadpoles of the Physalaemus cuvieri group and is thus characterized by an LTRF 2/3 with dorsal, ventrolateral and ventral gaps in the marginal papillae (Vera Candioti et al. 2011). During embryogenesis of the oral disc of Physalaemus, ventrolateral gaps appear in the marginal papillae, apparently in all species of the genus (see Vera Candioti et al. 2011). The ventrolateral gaps persist only in the tadpoles of Physalaemus cuvieri species group [except Physalaemus fischeri (Boulenger, 1890) and Physalaemus cicada Bokermann, 1966], in Physalaemus riograndensis Milstead, 1960 (Physalaemus henselii group) and in Physalaemus albifrons (see Vera Candioti et al. 2011). On the other hand, ventral gaps develop only in tadpoles of Physalaemus albifrons, in species of Physalaemus cuvieri group (except Physalaemus fischeri) and in two species of the Physalaemus henselii group [Physalaemus henselii (Peters, 1872) and Physalaemus fernandezae (Müller, 1926)]. Among the leiuperines, the ventrolateral gaps were only observed in some species of Pseudopaludicola (see Vera Candioti et al. 2011), and although its presence during development appears to be plesiomorphic for Physalaemus, its persistence in larval stages is a putative synapomorphy of the Physalaemus cuvieri group (including Physalaemus albifrons). Finally, the internal oral morphology of tadpoles of Physalaemus albifrons differs from that of Physalaemus biligonigerus, Physalaemus marmoratus and Physalaemus santafecinus based on the presence of three lingual papillae, which is a characteristic shared with some species of the Physalaemus cuvieri group (
Some of the species in the Physalaemus cuvieri group are sibling species with important intraspecific morphological variation. Therefore, the identification of these species that is based exclusively on their morphology is sometimes very difficult. Occasionally, species misidentification has occurred, for example, among Physalaemus cuvieri, Physalaemus albonotatus, Physalaemus cuqui and Physalaemus centralis (Barrio, 1965). Our results revealed conspicuous cytogenetic differences among most species of the Physalaemus cuvieri group. The exception is the great similarity between the karyotypes of Physalaemus albonotatus and Physalaemus cuqui. Additionally, the karyotypes of the species analyzed here were distinguished from the previously analyzed karyotype of Physalaemus cuvieri. The interspecific variation described in this work regarding heterochromatin and NOR distribution is of fundamental importance for the comparative analysis of the Physalaemus cuvieri species group.
An interstitial C-band was observed near the centromere in the long arm of chromosome 2 of Physalaemus albonotatus, Physalaemus centralis and Physalaemus cuqui; whereas in the karyotype of Physalaemus ephippifer (
Despite the overall similarity in chromosomal morphology among the species currently allocated to the Physalaemus cuvieri group, chromosome pairs 8 and 9 differ greatly. The differences in these chromosomes probably arose from the distinct locations of the NOR in these karyotypes, as these rDNA genes occupy different sites in pairs 8 and/or 9 of these species. The observed pattern of NOR occurrence can be helpful in distinguishing the analyzed species of the Physalaemus cuvieri group. Noticeably, a pericentromeric NOR site was found exclusively in the Physalaemus centralis karyotype. However, the NOR-bearing chromosomes (chromosome pairs 8) from the species Physalaemus cuvieri (
The authors are thankful to Dr. Denise de Cerqueira Rossa Feres for helping with field frog sampling and to Julián Faivovich and anonymous referees for revising the manuscript. This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (Fapesp) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). All specimens from Brazil were collected under a permit issued by the Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (IBAMA) (Proc. No. 02010.002895/03-84). DB is very grateful to the Instituto de Herpetología - Fundación Miguel Lillo, CONICET PIP 1112008010 2422, ANPCyT PICTs, 07–01485, 06–223, and 07–02202, and PICT-O 37035 for funding. DB, JMF, and CT are grateful to Comité Ejecutivo de Desarrollo e Innovación Tecnológica (CEDIT) of Misiones province. JMF thanks Carrera del Doctorado en Ciencias Biológicas de la FCEFyN, UNC.
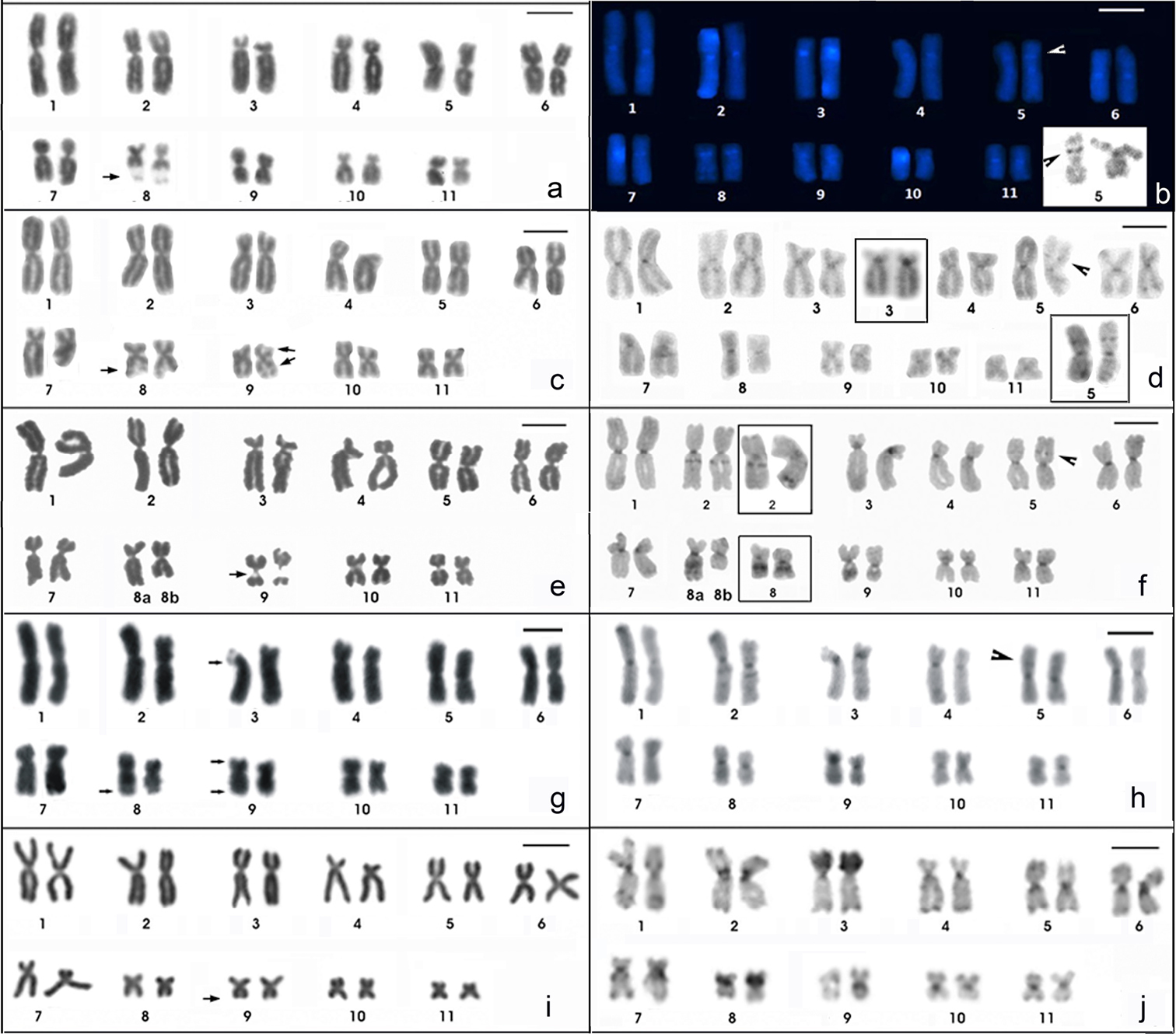
Giemsa-stained (a, c, e, g, i) and C-banded karyotypes of Physalaemus albifrons (a, b) Physalaemus albonotatus (c, d) Physalaemus centralis (e, f) Physalaemus cuqui (g, h) and Physalaemus santafecinus (i, j). In b the chromosomes were stained with DAPI after C-banding, except those in the inset, which were stained with Giemsa. In the insets in d C-banded chromosome pairs 3 and 5, showing evident pericentromeric and interstitial bands, respectively. The insets in f show the heteromorphic pair 2 and the homomorphic pair 8 of the ZUEC 13696 specimen. Arrows point NORs. Arrowheads point the C-band in 5p. Bar=10mm.