Citation: Kumbıçak Z, Ekiz E, Çiçekli S (2014) Karyotypes of six spider species belonging to the families Gnaphosidae, Salticidae, Thomisidae, and Zodariidae (Araneae) from Turkey. Comparative Cytogenetics 8(2): 93–101. doi: 10.3897/CompCytogen.v8i2.6065
In this study, the karyotypes of six spider species from Turkey belonging to the families Gnaphosidae, Salticidae, Thomisidae, and Zodariidae were analyzed. Male chromosomal features including diploid chromosome numbers and sex chromosome systems were determined as 2n=22, X1X20 in Drassyllus sur Tuneva & Esyunin, 2003, Nomisia exornata (C. L. Koch, 1839), and Nomisia orientalis Dalmas, 1921; 2n=28, X1X20 in Sitticus caricis (Westring, 1861); 2n=23, X0 in Xysticus gallicus Simon, 1875 and 2n=42, X1X20 in Pax islamita (Simon, 1873), respectively. The chromosome morphology of all species was acrocentric. Data obtained contribute to knowledge of the karyotype evolution of araneomorphs.
Araneae, diploid number, sex chromosome system
Spiders are one of the most important animal groups, and contain approximately 44 500 species all around the world (
Entelegyne spiders form a very diversified clade of araneomorphs. Their karyotypes are characterized by a predominance of acrocentric chromosomes, X1X20 sex chromosome system (
In spiders, the X1X20 system could be the ancestral sex chromosome determination as inferred from its presence in the most primitive recent spiders, namely the suborder Mesothelae and basal families of the infraorder Mygalomorphae (
Salticidae, Thomisidae, Gnaphosidae, and Zodariidae are some of the largest families in the order Araneae (
This study presents karyotypes of six species belonging to the genera Drassyllus Chamberlin, 1922 and Nomisia Dalmas, 1921 (Gnaphosidae), Sitticus Simon, 1901 (Salticidae), Xysticus C. L. Koch, 1835 (Thomisidae), and Pax Levy, 1990 (Zodariidae). Our study brings new data and fills some gaps in cytogenetics of these families.
Material: Spiders were collected in Mediterranean, Southeast and Central Anatolia (Turkey) during the year 2012. Collection data of particular species (localities including their coordinates, dates of collection, number of individuals studied) are listed in Table 1. Voucher specimens were deposited in the collection of Department of Molecular Biology and Genetics, Art and Science Faculty, Nevşehir Hacı Bektaş Veli University (Nevşehir, Turkey). The identification of spiders was made by O. Seyyar (Department of Biology, Art and Science Faculty, Niğde University, Niğde, Turkey).
Table 1. Material used for chromosome analysis.
| Family | Species | Locality | Coordinates | Date of Collection | Number of Individuals Studied |
| Gnaphosidae | Drassyllus sur Tuneva & Esyunin, 2003 | Gaziantep, Sakçagözü | 37°10'18"N, 36°55'39"E | 04.04.2012 | 7♂ |
| Nomisia exornata (C. L. Koch, 1839) | Antalya, Aksu | 36°55'30"N, 30°48'29"E | 24.03.2012 | 11♂ | |
| Nomisia orientalis Dalmas, 1921 | Antalya, Gazipaşa | 36°16'23"N, 32°17'33"E | 24.03.2012 | 2♂ | |
| Osmaniye, Düziçi | 37°15'02"N, 36°26'36"E | 21.05.2012 | 5♂ | ||
| Adıyaman, Kahta | 37°48'46"N, 38°38'20"E | 11.03.2012 | 4♂ | ||
| Gaziantep, Islahiye | 37°01'21"N, 36°37'24"E | 06.04.2012 | 9♂ | ||
| Salticidae | Sitticus caricis (Westring, 1861) | Nevşehir, Göreme | 38°38'44"N, 34°50'06"E | 10.05.2012 | 8♂ |
| Nevşehir, Zelve | 38°40'16"N, 34°51'43"E | 27.06.2012 | 3♂ | ||
| Thomisidae | Xysticus gallicus Simon, 1875 | Adana, Çamalan | 37°19'12"N, 34°36'28"E | 12.04.2012 | 6♂ |
| Mersin, Bozyazı | 36°06'04"N, 32°58'38"E | 15.04.2012 | 2♂ | ||
| Mersin, Aydıncık | 36°08'36"N, 33°22'59"E | 15.04.2012 | 3♂ | ||
| Zodariidae | Pax islamita (Simon, 1873) | Osmaniye, Toprakkale | 37°04'24"N, 36°08'42"E | 09.06.2012 | 5♂ |
Chromosome preparations and observation: Slides for chromosome observations were made by the spreading technique of
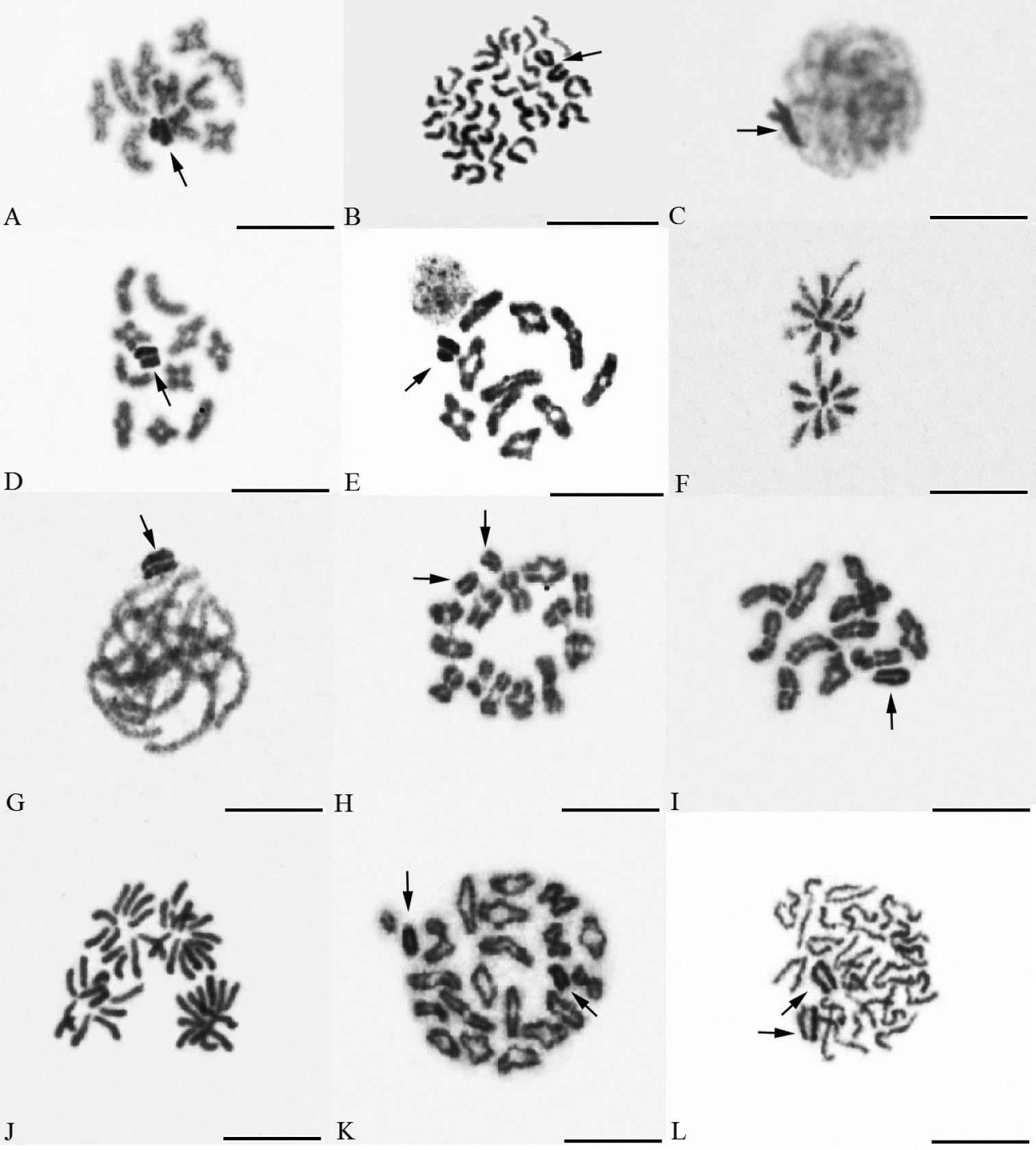
The chromosomes of Drassyllus sur Tuneva & Esyunin, 2003 (2n♂=22) were acrocentric. The sex chromosome system was formed by chromosomes X1 and X2 which were medium-sized elements (Fig. 1A). The autosome pairs decreased gradually in size. Length of autosome pairs decreased from 9.74±0.29% to 6.89±0.12% of TCL. Relative length of X1 and X2 was 8.45±0.06% and 7.57±0.17% of the diploid set, respectively.
Karyotypes of species based on spermatogonial metaphases. A Drassyllus sur, 2n♂=22, X1X20 B Nomisia exornata, 2n♂=22, X1X20 C Nomisia orientalis, 2n♂=22, X1X20 D Sitticus caricis, 2n♂=28, X1X20 E Xysticus gallicus, 2n♂=23, X0 F Pax islamita 2n♂=42, X1X20. Bar=10 µm.
There were 10 autosomal bivalents and two sex chromosomes at diplotene (Fig. 2A). Sex chromosomes were positively heteropycnotic from leptotene to metaphase II (Fig. 2B).
Meiosis of gnaphosid, salticid, thomisid and zodariid males. Drassyllus sur A diplotene B metaphase II, Nomisia exornata C early pachytene D diakinesis, Nomisia orientalis E diakinesis F part of anaphase II showing one plate with 10 chromosomes and another plate with 12 chromosomes, Sitticus caricis G pachytene H diplotene, Xysticus gallicus I diakinesis J anaphase II, Pax islamita K diakinesis L half of metaphase II (arrows indicate sex chromosomes). Bar=10 µm.
The karyotype of Nomisia exornata (C. L. Koch, 1839) (Fig. 1B) (2n♂=22, X1X20) was acrocentric. Autosome pairs decreased gradually in size from 10.3±0.21% to 5.85±0.17% of TCL. Relative length of X1 and X2 were 7.46±0.13% and 6.65±0.08% of TCL, respectively.
The autosomes of Nomisia orientalis Dalmas, 1921 (Fig. 1C) (2n♂=22, X1X20) was acrocentric. RCL of autosome pairs were decreased gradually from 10.61±0.24% to 6.62±0.19% of TCL. The gonosomes X1 (7.91±0.12% of TCL) and X2 (6.10±0.07% of TCL) showed acrocentric morphology.
The sex chromosomes were positively heteropycnotic from leptotene to diakinesis in both Nomisia species studied. Plates consisted of 10 autosomal bivalents and two univalent sex chromosomes from pachytene to metaphase I (Fig. 2C–E). At meiotic anaphases, 10 chromosomes segregated to one pole and 12 chromosomes to another pole (Fig. 2F).
The autosomes of Sitticus caricis (Westring, 1861) (2n♂=28, X1X20) were acrocentric. RCL decreased gradually from 8.47±0.42% to 5.04±0.16% of TCL (Fig. 1D). The sex chromosomes X1 (7.33 ±0.51% of TCL) and X2 (6.72±0.38% of TCL) were medium sized in comparison with the autosomes.
Leptotene, zygotene, and pachytene nuclei included a positively heteropycnotic sex chromosome body that was located at the periphery of the nucleus (Fig. 2G). At late prophase I (i.e. diplotene and diakinesis), 13 autosomal bivalents and two univalent sex chromosomes were determined (Fig. 2H).
The chromosome set of Xysticus gallicus Simon, 1875 (2n♂=23, X0) contained 11 acrocentric pairs and a small X chromosome (Fig. 1E). Autosome pairs decreased gradually in size from 10.28±0.62% to 6.46±0.39% of TCL. Relative length of X chromosome was 6.77±0.46% of TCL. This chromosome was longer than the smallest autosome pair.
From leptotene to diakinesis, X chromosome was formed by an intensively stained material. Diakinetic plates exhibit 11 autosomal bivalents (Fig. 2I). At metaphase II and anaphase II, X chromosome was isopycnotic with autosomes (Fig. 2J).
The karyotype of Pax islamita (Simon, 1873) consisted of acrocentric chromosomes; the diploid number was 42 (Fig. 1F). Autosome pairs exhibited a gradual decrease of relative lengths from 6.42±0.58 to 3.31±0.24% of TCL. This species showed X1X20 sex chromosome system. The acrocentric gonosomes showed similar size. Their relative lengths were 5.92±0.66% and 5.37±0.18% of TCL, respectively.
From beginning of pachytene to metaphase I, plates consisted of 20 autosomal bivalents and two not associated sex chromosomes on the periphery of nucleus (Fig. 2K). Sex chromosomes were positively heteropycnotic during prophase and metaphase II. Metaphases II consisted of 20 or 22 chromosomes, respectively. Metaphases II with 22 chromosomes contained two X chromosomes (Fig. 2L).
Karyotypes of 771 spider species from 277 genera are known at present (
So far, chromosome numbers have been established for 38 species of gnaphosid spiders. The majority of species (33 in a total) have 2n♂=22 including X1X20 sex chromosome system (
Male diploid numbers in salticids vary from 2n=14 in Menemerus illigeri (Audouin, 1826) (
The male karyotype of Xysticus gallicus displays the general pattern described for most Thomisidae: a diploid chromosome number 23 and X0 sex chromosome system including acrocentric sex chromosome. All Xysticus species analyzed so far present this karyotype with exception of Xysticus triguttatus Keyserling 1880 (
Our study represented a diploid number of 42 acrocentric chromosomes and X1X20 system in Pax islamita. This finding is compatible with the results reported by
In conclusion, our study described the karyotype features of five araneomorph spiders for the first time and confirms some findings of
We are very thankful to four anonymous referees for valuable suggestions and improvements of the manuscript. This study was carried out in the Department of Molecular Biology and Genetics, Faculty of Science and Art, Nevşehir Hacı Bektaş Veli University (Nevşehir, Turkey) and supported by Nevşehir Hacı Bektaş Veli University Directorate of Scientific Research Projects (NEUBAP13F10).