Citation: Spoz A, Boron A, Porycka K, Karolewska M, Ito D, Abe S, Kirtiklis L, Juchno D (2014) Molecular cytogenetic analysis of the crucian carp, Carassius carassius (Linnaeus, 1758) (Teleostei, Cyprinidae), using chromosome staining and fluorescence in situ hybridisation with rDNA probes. Comparative Cytogenetics 8(3): 233–248. doi: 10.3897/CompCytogen.v8i3.7718
The crucian carp Carassius carassius (Linnaeus, 1758) is a species with restricted and decreasing distribution in Europe. Six males and six females of the species from the Baltic Sea basin in Poland were examined to show sequentially CMA3/AgNO3 staining pattern, DAPI staining, and, for the first time in literature, molecular cytogenetic analysis using double-colour fluorescence in situ hybridisation (FISH) with 28S and 5S rDNA probes. The karyotype consisted of 20 m, 36 sm and 44 sta chromosomes, NF=156. The AgNO3 stained NORs were most frequently located terminally in the short arms of two sm and two sta elements, and CMA3-positive sites were also observed suggesting abundant GC-rich repetitive DNA in the regions. Other CMA3-positive sites in the short arms of six to ten sm and sta chromosomes were detected. The results based on 28S rDNA FISH confirmed the location of rDNA sites. DAPI-negative staining of NORs suggested the scarcity of AT-rich DNA in the regions. FISH with 5S rDNA probe revealed 8–14 loci (ten and 12 in respectively 49 and 29% of metaphases). They were located in two sm and eight to ten sta chromosomes and six of them were larger than others. Simultaneously, mapping of the two rDNA families on the chromosomes of C. carassius revealed that both 28S and 5S rDNA probes were located in different chromosomes. Molecular cytogenetic data of C. carassius presented here for the first time give an important insight into the structure of chromosomes of this polyploid and declining species and may be useful in its systematics.
Cyprinidae, CMA3, FISH with rDNA, molecular cytogenetics, NOR-phenotype, polyploid species
The genus Carassius Jarocki, 1882 is a fish group of polyploid origin as are some other cyprinids of subfamilies Cyprininae and Barbinae s.l., e.g. Cyprinus Linnaeus, 1758 and Barbus Cuvier, 1816 (
The crucian carp, Carassius carassius (Linnaeus, 1758), native to Europe, is widely distributed from the northern France to the Danube drainage and Siberia, and from England in the north to the Alps in the south. This species is adapted to both a wide range of temperature and low oxygen content and prefers densely vegetated water bodies−backwaters and oxbows of lowland rivers, and lakes (
The crucian carp is included in the least concern IUCN category but is regarded as disappearing in many water bodies of its range (
The karyotype of this species has been described by
The location of ribosomal genes in the chromosomes is commonly used as very informative cytogenetic features (
In the present study, the crucian carp Carassius carassius was examined for the chromosomal distribution of the nucleolar organiser regions (NORs) using sequential staining with silver nitrate (AgNO3), chromomycin A3 (CMA3), and DAPI staining. Moreover, fluorescence in situ hybridisation (FISH) with 28S (major) and 5S (minor) rDNA probes was performed. This is the first report of simultaneous localisation of two rDNA families (45S and 5S rDNA) in chromosomes of Carassius carassius. The ribosomal gene distribution data extend our knowledge on the cytotaxonomy and gave us information about functional structure of the chromosomes in this polyploid and declining species.
In total 12 individuals, six males and six females of Carassius carassius (of the average length and body weight respectively, 165.0 mm and 140.0 g for females, and 151.0 mm and 124.0 g for males) were studied. They were collected from the Kortowskie Lake (53°45'43"N, 20°26'42"E), the Pregola River drainage (Baltic Sea basin) by net and then transported alive to the laboratory. Species identification followed
Mitotic chromosome preparations were made from each individual following
Chromosomes were stained with a solution of 4% Giemsa (pH=6.8) and then classified according to
Chromosome slides of three males and three females were sequentially stained with AgNO3 and CMA3 according to
Single colour FISH with human 28S rDNA probe or double-colour FISH with loach 5S and human 28S rDNA probes were used according to
Hybridisation signals in at least 15 metaphase figs of each individual were observed under a Nikon Eclipse E800 fluorescence microscope using a Nikon B-2A filter for a single colour FISH and black and white CCD camera Pixera Penguin 150CL-CU (Pixera), and a Nikon Eclipse 90i fluorescence microscope equipped with ProgRes MFcool camera (Jenoptic) for capturing the images of a dual colour FISH. The images were processed using Penguin Mate ver. 1.0.8. software for RGB pseudocolour imaging (Pixera) and Lucia ver. 2.0 (Laboratory Imaging).
Voucher specimens were preserved frozen and deposited at the Department of Zoology, University of Warmia and Mazury in Olsztyn, Poland.
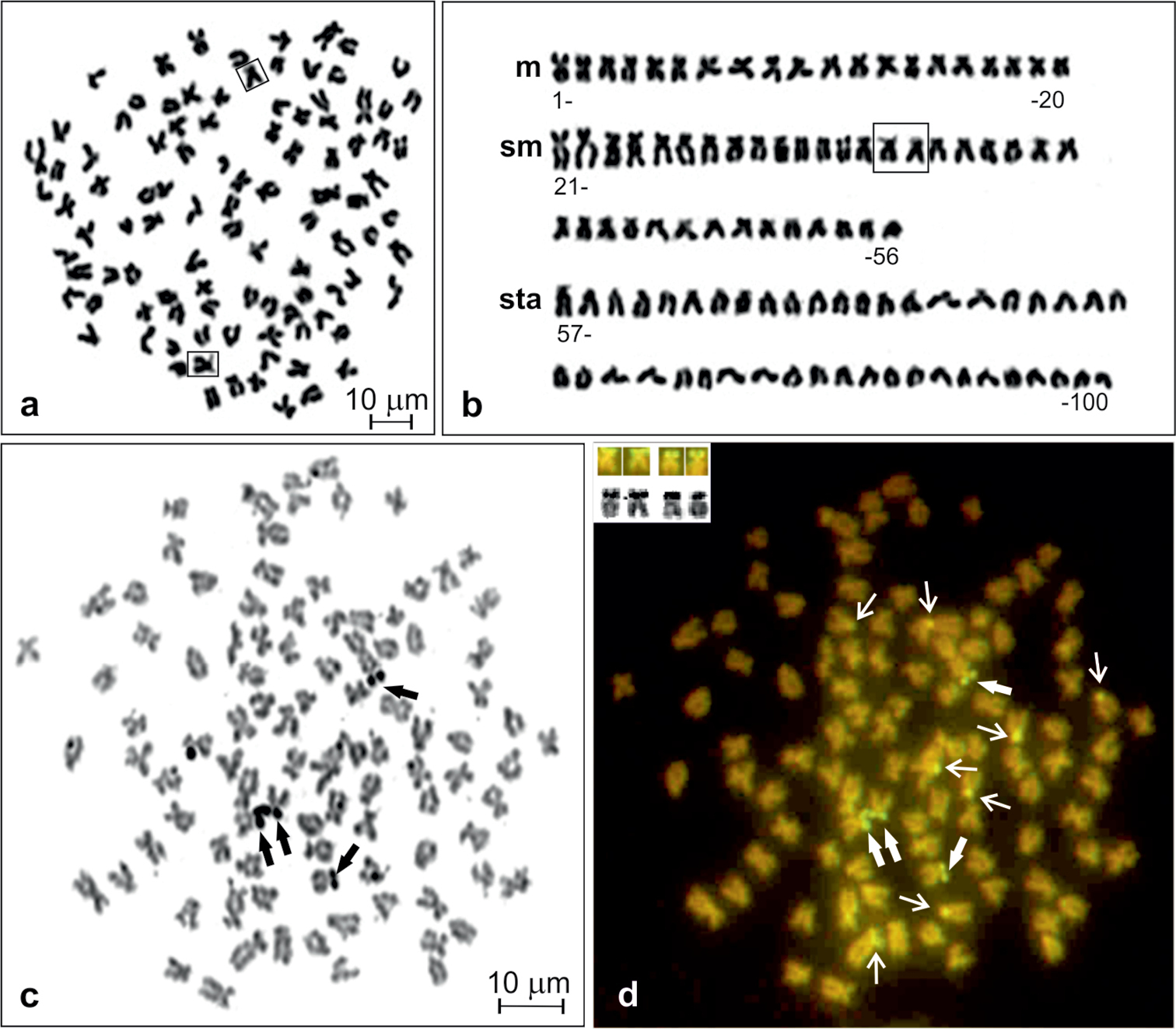
The crucian carp from the Kortowskie Lake exhibits a diploid chromosome number of 100 (Fig. 1a) without any supernumerary chromosomes in 369 (94.4%) out of 391 analysed metaphase figs. The karyotype consisted of 20 m, 36 sm and 44 sta chromosomes (Fig. 1b). The chromosome arm number (NF) was counted as 156. The first submetacentric pair (11th pair) was easily recognisable in all metaphase figs, being the largest elements in the chromosome complement. No variability in the chromosome formula was observed and heteromorphic sex chromosomes were not detected.
Giemsa stained metaphase (a), corresponding karyotype of Carassius carassius (b), and metaphase spread sequentially stained with AgNO3 (c) and CMA3 (d). NOR chromosomes shown in frames (in a and b), Ag-NORs and corresponding CMA3-positive sites shown by thick arrows (in c and d) and shown in inset (in d), other CMA3–positive sites shown by thin arrows (in d).
AgNO3 stained active nucleolus organiser regions (AgNORs) were located terminally at the short arms of two sm and two st chromosomes (Fig. 1c). After sequential staining with CMA3, all signals were observed as a distinct bright fluorescence, suggesting abundant GC-rich repetitive DNA sequences in the regions (Fig. 1d). Among 90 metaphases of six individuals, two to four CMA3-positive sites corresponding to AgNORs were detected, but 57.8% of metaphases showed four bright signals. Three and two such sites were observed, respectively, in 31.1 and 11.1% of analysed metaphases. In addition, there were extra CMA3-positive sites located at the short arms of six to ten sm and sta chromosomes. Most frequently (in 52.2% of metaphases) eight such sites at four of each of sm and sta elements (Fig. 1d) or six (in 32.2% of metaphases) sites at three of each of sm and sta were observed.
One of the submetacentric chromosomes (chromosome no. 35 of pair 18, shown in frame in Fig. 1a–b), possessing clearly visible secondary structure along its short arm, was easily distinguishable among others in all metaphase figs stained with Giemsa.
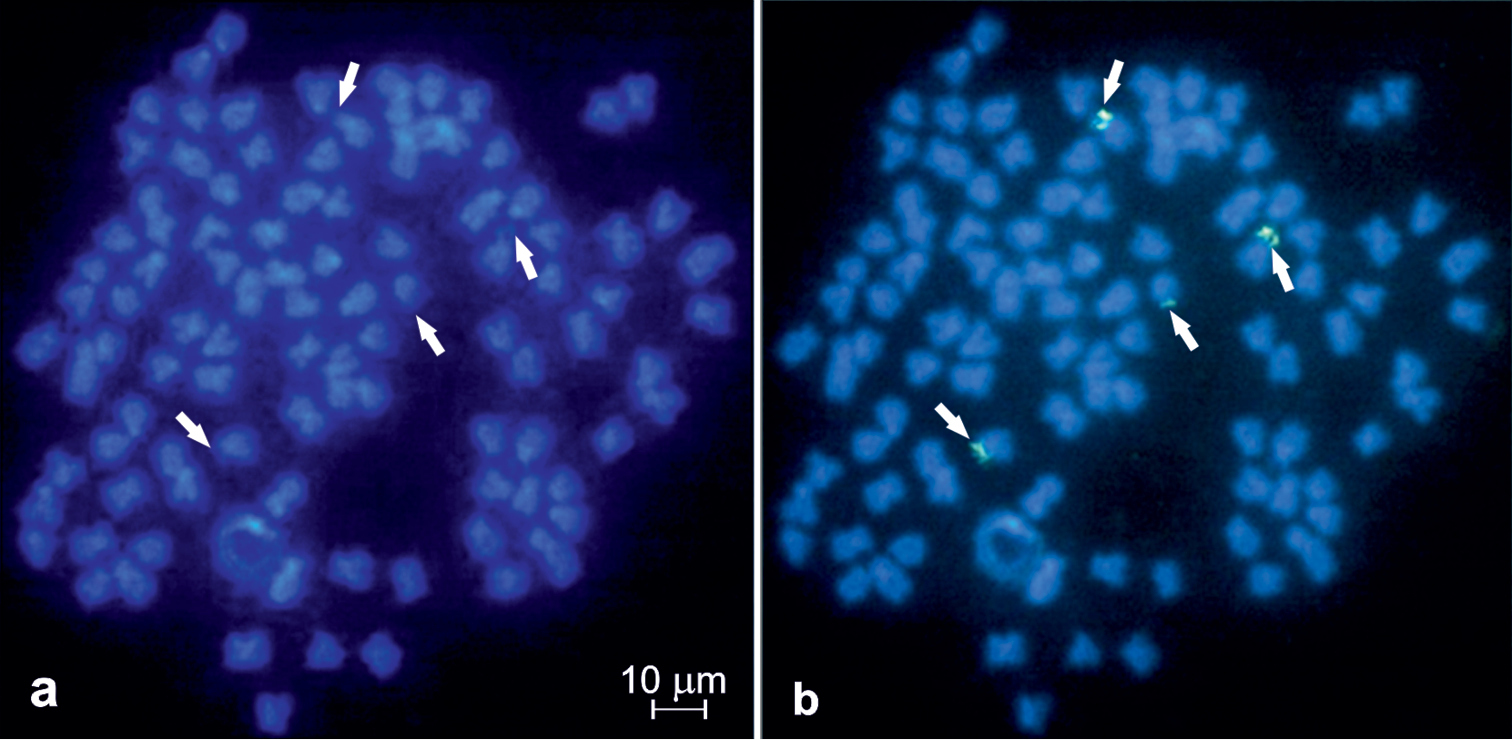
DAPI-counterstained chromosomes have shown some slightly visible AT-rich pericentromeric heterochromatic regions of 12–14 sta and at the short arms of four to six sm (Fig. 2a–b). However, they were not detected in the metaphase figs after using dual colour FISH that such the chromosomal regions were dimly DAPI-stained (Fig. 3a).
Metaphase fig of Carassius carassius DAPI stained (a) and with a single colour FISH (b) with 28S rDNA probe. 28S rDNA hybridisation signals shown by arrows.
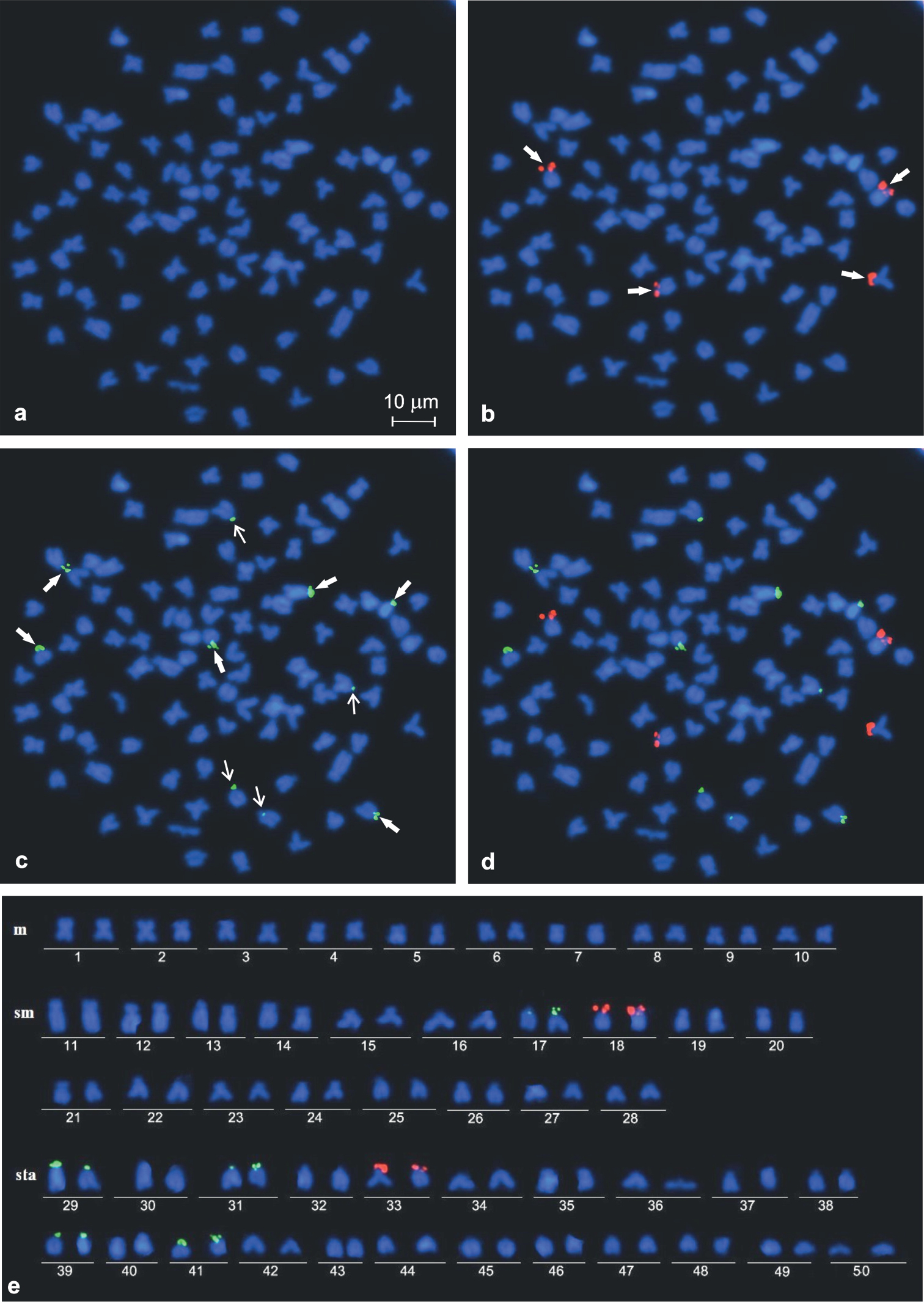
Representative mitotic metaphase figs (a−d) and corresponding karyotype of Carassius carassius (e): a DAPI stained and b−d most frequent hybridisation pattern after dual colour FISH with four 28S rDNA sites (b), ten 5S rDNA sites (c) and both rDNA probes (d). Six stronger and four weaker 5S rDNA hybridisation sites (c) shown by thick and thin arrows, respectively.
Single FISH using 28S rDNA probe analysed in 68 metaphase figs of two females and two males and dual colour FISH analysed in 243 metaphase figs of four females and four males revealed either three or four loci in their chromosome complement. In most of the metaphase figs (76.2%), the signals were found in the short arms of two each of sm and st chromosomes (Figs 2b, 3b). Three hybridisation sites were observed commonly as intense and large signals, whereas the signal in the fourth site was smaller and weaker than the other three sites. DAPI-negative staining of the observed NORs suggested the scarcity of AT-rich DNA in the regions (Figs 2a, b, 3a, b). In the rest of the analysed metaphase figs (23.8%), three 28S rDNA sites were observed in the short arms of two sm and one sta elements. Numerous metaphases showed close association of NORs involving two or sometimes three chromosomes.
FISH with 5S rDNA probe analysed in 243 metaphase figs of four males and four females revealed an unexpectedly large number of loci, from eight to 14. The obtained hybridisation signals had different intensities on various chromosomes and could be classified as strong and weak (Fig. 3c–d). All individuals frequently showed 10 (Fig. 1c) or 12 such loci in respectively 48.6% and 29.2% of metaphase figs. They were located at the short arms of two sms (pair 17 in Fig. 3e) and at the short arms or in a subcentromeric position of eight to ten sta chromosomes (pairs 29, 31, 39 and 41 in Fig. 3e). Six hybridisation sites of 5S rDNA were stronger than the other four to six (Fig. 3c–e). Among 15.2% and 7.0% of the rest of metaphase figs, the 5S rDNA loci were located, respectively, in eight and 14 chromosomes. Usually, in metaphase figs containing 14 signals, two signals were very weak.
Thus, Carassius carassius was characterised by the modal number of ten 5S rDNA loci. Signal heteromorphism was detected on the homologous chromosome of pairs 17 and 31 (Fig. 3e). Both classes of rDNA probes were always located in different chromosomes and co-localisation in the same chromosome was not observed (Fig. 3d).
No sex-dependent variability in the cytogenetic features was found.
Undoubtedly, the crucian carp Carassius carassius possesses 2n=100 chromosomes in its somatic cells but data on the karyotype reported in literature somewhat differ (Table 1). The reason for this could be that the karyotype of the crucian carp contains a lot of very small chromosomes which are similar in size. The problem mainly concerns discrimination between sm and sta chromosomes as it occurs in the karyotype of a related species Carassius gibelio (
Cytogenetical data of the crucian carp, Carassius carassius. Symbols of chromosomes: m – metacentric, sm – submetacentric, sta – subtelo- to acrocentric, NF – number of chromosome arms.
| L.p | Locality | 2n | Karyotype | NF | Cytogenetic features | Reference |
|---|---|---|---|---|---|---|
| 1. | - | 94 | - | - | - | |
| 2. | - | 104 | 20m+72sm+12a | 196 | - | |
| 3. | the Netherlands (Baltic basin) | 100 | 20m+40sm+40a | 160 | - | |
| 4. | France (Garonne drainage) | 100 | 20m+44sm+36a | 164 | - | |
| 5. | Drina R. (Danube), Bosnia | 100 | 52m, sm+48sta | 152 | - | |
| 6. | Danube R., Romania | 50 | 20m+12sm+18sta | 82 | - | |
| 7. | Water bodies in Moscow region, Russia (Volga drainage) | 100 | 48m, sm+52sta | 148 | - | |
| 8. | Elbe R., Czech Republic | 100 | - | - | AgNOR | |
| 9. | the Netherlands (Baltic basin) | 100 | 20m+40sm +40a | 160 | - | |
| 10. | Tarim R., Xinjiang, China | 100 | 32m+34sm+34sta | 166 | - | |
| 11. | Elbe R., Czech Republic | 100 | 20m+36sm+44sta | 156 | C bands, AgNOR, DAPI/CMA3 | |
| 12. | Kortowskie Lake, Pregola R. drainage, Poland | 100 | 20m+36sm+44sta | 156 | AgNOR/CMA3, 45S and 5S rDNA (FISH) | present study |
The crucian carp and other Carassius species distributed in Europe were recognised as monophyletic lineages (
Most of the cyprinid species, for example those from the subfamilies Leuciscinae, Gobioninae and Danioninae, are characterised by 2n=50 or 2n=48 chromosomes (e.g.
The number of four AgNORs (two sm and two st) characterises the karyotype of Carassius carassius (
We documented that the AgNOR sites were CMA3 positive similar to what is found in many other Teleostei (
The karyotype of Carassius carassius after DAPI staining described by
The results from FISH with 28S rDNA confirmed for the first time in literature that the karyotype of Carassius carassius (2n=100) is characterised by the conservative number of NORs − four − located in the short arms of two sm and two st chromosomes. It was mentioned by
The weak or missing signal of hybridisation in one out of the four NORs in the karyotype of Carassius carassius could be due to either a low copy number of 28S rDNA or a deletion of these genes, or due to technical reasons (
The FISH localisation of the 5S rDNA revealed that these sequences are spread in at least eight chromosomes. Carassius auratus (2n=100) is characterised by 5S rDNA large hybridisation sites located at the short arms of two st and from two to eight smaller 5S rDNA sites whereas a triploid form of Carassius gibelio (3n=162) had three larger sites and from six to 18 small ones (
Commonly in Teleostei, there is a single locus for the 5S ribosomal sequences, which is regarded as an ancestral condition while the hybridisation pattern with two or more loci may be considered as a derived state (
The 5S rDNA clusters in fishes seem to be most frequently located at interstitial chromosome sites as they were found in most fish species in different orders (
In most of the described fish species including cyprinids, the two rDNA families are located at different chromosomes (
We updated the information on the karyotype, showed for the first time sequentially CMA3/AgNO3 banding pattern and also provided new molecular cytogenetic data on the crucian carp Carassius carassius using double-colour FISH with 5S and 28S rDNA probes. The obtained results improve our knowledge about the chromosome structure and physical location of major and minor ribosomal sequences in this fish species. Moreover, the results herein gave an important insight into the molecular cytotaxonomy of the crucian carp a polyploid and declining species and may be useful in its systematics.
We would like to express our sincere thanks to Dr Slawomir Boron for collecting fish. This work was carried out partially within the grant no. 2P04C 08229 supported financially by the State Committee for Scientific Research KBN and the project no. 0208.806 of the University of Warmia and Mazury in Olsztyn, Poland. A part of this study was also supported by the JSPS Invitation Fellowship for Research in Japan (S-05135) to Alicja Boron. We are grateful to two anonymous reviewers for their comments which helped to improve the manuscript.