Citation: Hodaňová L, Kalous L, Musilová Z (2014) Comparative cytogenetics of Neotropical cichlid fishes (Nannacara, Ivanacara and Cleithracara) indicates evolutionary reduction of diploid chromosome numbers. Comparative Cytogenetics 8(3): 169–183. doi: 10.3897/CompCytogen.v8i3.7279
A comparative cytogenetic analysis was carried out in five species of a monophyletic clade of neotropical Cichlasomatine cichlids, namely Cleithracara maronii Steindachner, 1881, Ivanacara adoketa (Kullander & Prada-Pedreros, 1993), Nannacara anomala Regan, 1905, N. aureocephalus Allgayer, 1983 and N. taenia Regan, 1912. Karyotypes and other chromosomal characteristics were revealed by CDD banding and mapped onto the phylogenetic hypothesis based on molecular analyses of four genes, namely cyt b, 16S rRNA, S7 and RAG1. The diploid numbers of chromosomes ranged from 44 to 50, karyotypes were composed predominantly of monoarmed chromosomes and one to three pairs of CMA3 signal were observed. The results showed evolutionary reduction in this monophyletic clade and the cytogenetic mechanisms (fissions/fusions) were hypothesized and discussed.
Cichlid cytotaxonomy, cyt b, 16S rRNA, S7-1, RAG1 phylogeny, karyotype differentiation, CMA3 phenotypes , Cichlasomatini
Cichlids are a species-rich group of ray-finned fishes (Actinopterygii), distributed in tropical and subtropical freshwaters of Africa and South and Central America, Texas, Madagascar, the Middle East, India and Sri Lanka (
In general, genomes of ray-finned fishes are known for high evolutionary dynamics among vertebrates, which is reflected in huge genome-architecture variability (
In total, over 190 cichlid species have been cytogenetically analyzed and the karyotype formula was determined for 157 of them (
In the past only few species were analyzed and Neotropical cichlids were considered a karyotypically conservative group due to the frequent findings of 48 chromosomes (
Dwarf cichlids of the genus Nannacara Regan, 1905, and its relatives, genera Ivanacara Römer & Hahn, 2007 and Cleithracara Kullander & Nijssen, 1989 represent a well-defined evolutionary lineage of acaras (NIC-clade of the tribe Cichlasomatini,
In this study we present karyotypes and other chromosomal characteristics as revealed by CDD banding in five species of monophyletic clade of neotropical Cichlasomatine cichlids, namely Cleithracara maronii, Ivanacara adoketa (Kullander & Prada-Pedreros, 1993), Nannacara anomala, Nannacara aureocephalus Allgayer, 1983 and Nannacara taenia Regan, 1912. We further mapped the results onto the phylogenetic hypothesis from molecular analyses based on four genes. We discuss possible scenario of the karyotype evolution of the clade of dwarf cichlids within the tribe Cichlasomatini.
The species included in the present study are listed in Table 1. Most of the individuals originated from aquarium trade from different breeders. Further, various collectors or ornamental-fish importers donated several samples for DNA analysis. Species were identified following
Chromosomes were obtained by non-destructive isolation procedure from caudal fin regenerates as developed by
DNA was extracted from the ethanol-preserved samples by the commercially available kits (QiaGen), and four target genes (cyt b, 16S rRNA, S7 first intron, RAG1) were amplified by PCR using primers according to
Sample list for the present study. Details on individuals of cichlids investigated for the molecular genetics. Outgroup data were used from the original study (
| Individuals used in molecular phylogenetic analyses: | Accesion numbers in GenBank | Sample voucher | |||||
|---|---|---|---|---|---|---|---|
| species | sample code | origin | cytb | 16SrRNA | S7 | RAG1 | |
| Geophagus brasiliensis | outgroup – used from GenBank | EF470895 | EU888080 | EU199082 | EU706360 | - | |
| Bujurquina vittata | outgroup – used from GenBank | EF432951 | EF432892 | EF432984 | EU706385 | - | |
| Aequidens metae | outgroup – used from GenBank | EF432927 | EF432882 | EF432974 | - | - | |
| Laetacara thayeri | outgroup – used from GenBank | AY050608 | EF432909 | EF433001 | EU706401 | - | |
| Cleithracara maronii | Cleith | aquarium trade | AY050614 | EF432901 | EF432993 | EU706394 | ICCU 0736 |
| Nannacara (Ivanacara) adoketa | ADO | aquarium trade | EF432946 | EF432903 | EF432995 | EU706396 | ICCU 0745 |
| Nannacara (Ivanacara) adoketa | In06 | Rio Inirida | KJ136667 | - | KJ136659 | - | ICCU 1001 |
| Nannacara (Ivanacara) adoketa | In03 | Rio Inirida | KJ136668 | - | KJ136660 | - | ICCU 1002 |
| Nannacara anomala | ANO | aquarium trade | AY050618 | EF432898 | EF432990 | EU706391 | ICCU 0746 |
| Nannacara anomala | NaD | Orinoco delta | KJ136669 | KJ136671 | KJ136661 | - | ICCU 1004 |
| Nannacara anomala "Suriname" | WSN | F1 progeny | - | - | KJ136654 | - | - |
| Nannacara aureocephalus "blue" | RNA01 | aquarium trade | - | KJ136673 | KJ136663 | - | ICCU 1005 |
| Nannacara aureocephalus "blue" | RNA03 | aquarium trade | - | KJ136674 | KJ136664 | - | - |
| Nannacara aureocephalus "blue" | RNA04 | aquarium trade | - | KJ136675 | KJ136665 | - | - |
| Nannacara aureocephalus | AUR | aquarium trade | EF432939 | EF432899 | EF432991 | EU706392 | ICCU 0747 |
| Nannacara sp. | SAR | import/unknown | - | KJ136670 | KJ136655 | KJ136666 | ICCU 1003 |
| Nannacara prope aureocephalus "brown" | AurBrown01 | aquarium trade | - | KJ136672 | KJ136662 | - | - |
| Nannacara sp. "Soumourou" | NSP01 | F1 progeny | - | - | KJ136656 | - | - |
| Nannacara sp. "Oyapock" | NSP02 | F1 progeny | - | - | KJ136657 | - | - |
| Nannacara sp. "Oyapock" | NSP03 | F1 progeny | - | - | KJ136658 | - | - |
| Nannacara sp. | AF045860 | GenBank | - | AF045860 | - | - | - |
| Nannacara taenia | TAE | aquarium trade | EF432921 | EF432900 | EF432921 | EU706393 | ICCU 0749 |
Sample list for karyotypes analysis.
| Individuals used in cytogenetic analyses (all from aquarium trade): | ||
|---|---|---|
| Species | Number of analyzed individuals | Sex |
| Cleithracara maronii | 3 | undifferentiated |
| Ivanacara adoketa | 3 | 2× male, 1× female |
| Nannacara anomala | 5 | 3× male, 2× female |
| Nannacara aureocephalus | 3 | undiferentiated |
| Nannacara taenia | 3 | undiferentiated |
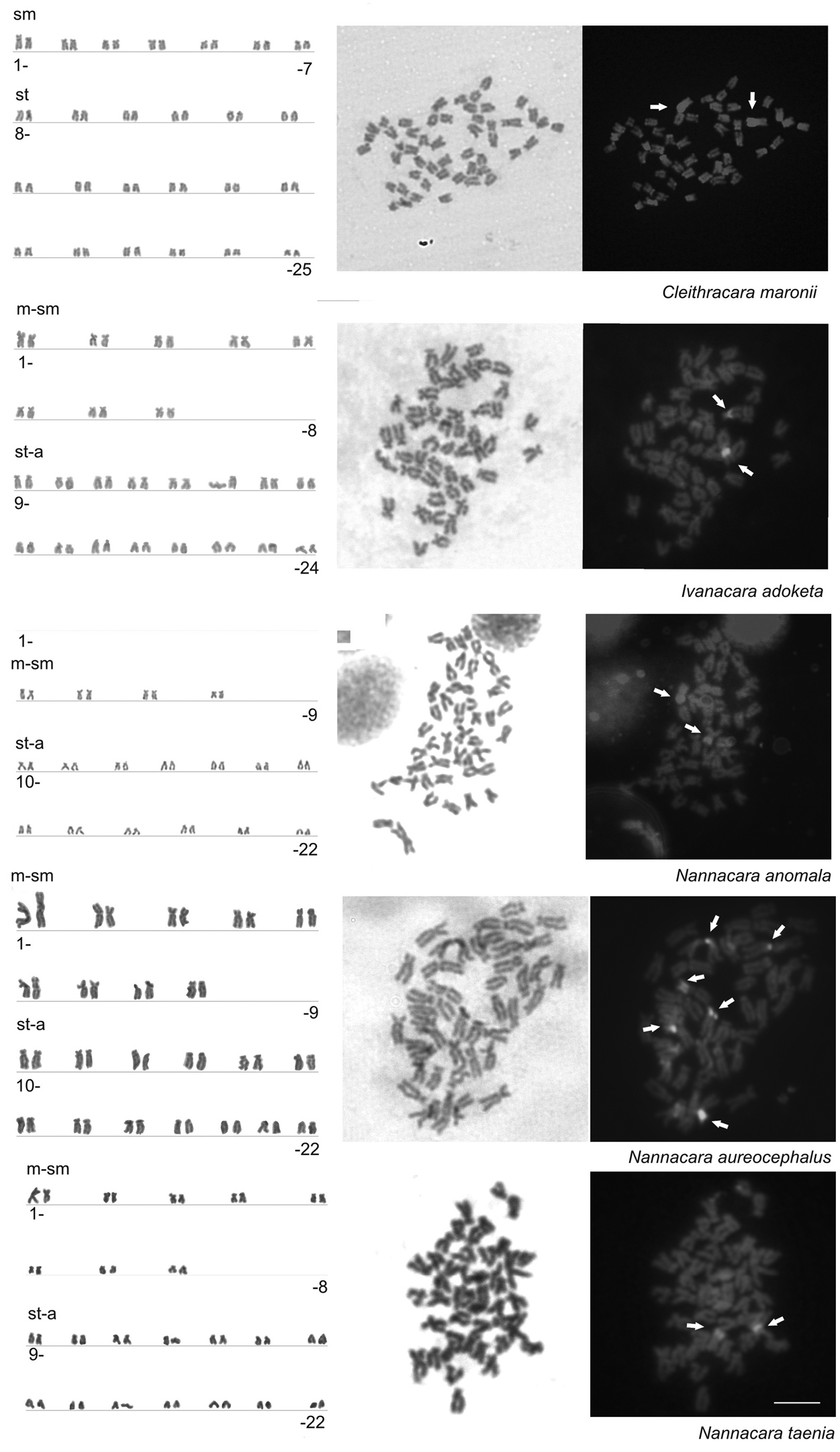
Results are summarized in Fig. 1 and Table 3. Examined individuals of the species of genera Nannacara, Ivanacara and Cleithracara showed the diploid chromosome number 2n = 44 to 50 chromosomes. All three species of the genus Nannacara possessed 44 chromosomes and karyotype composed of 18 metacentric (m)-submetacentric (sm)+26 subtelocentric (st)-acrocentric (a) or 16m-sm+28st-a chromosomes, while Ivanacara adoketa had 2n = 48 and karyotype of 16m-sm+32st-a chromosomes, and Cleihtracara maronii had 2n = 50 composed of 14sm+36st-a chromosomes. Karyotypes of all studied species are shown in Fig. 1.
In the karyotypes of four studied species, namely Cleithracara maronii, Ivanacara adoketa, Nannacara anomala, and Nannacara taenia, the CMA3-positive signals were found on one chromosome pair, although probably not homologous in different species. In Cleithracara maronii the CMA3-positive signals were located on terminal parts of the largest m-sm chromosome pair, whereas in Ivanacara adoketa and Nannacara taenia the CMA3 signals were located a chromosome pair from st-a group, terminal parts in Nannacara taenia and around the centromere in Ivanacara adoketa. In Nannacara anomala the CMA3 signals were found on the terminal parts of a chromosome pair from m-sm group, but not on the largest pair. Contrarily, in the karyotype of Nannacara aureocephalus, the CMA3 signals were located on three m-sm chromosome pairs including the largest chromosome pair in the centromeric region. See Table 3 for more detail about the karyotype formulas and CMA3 phenotypes and Fig. 1 for representative metaphases and results of different staining steps.
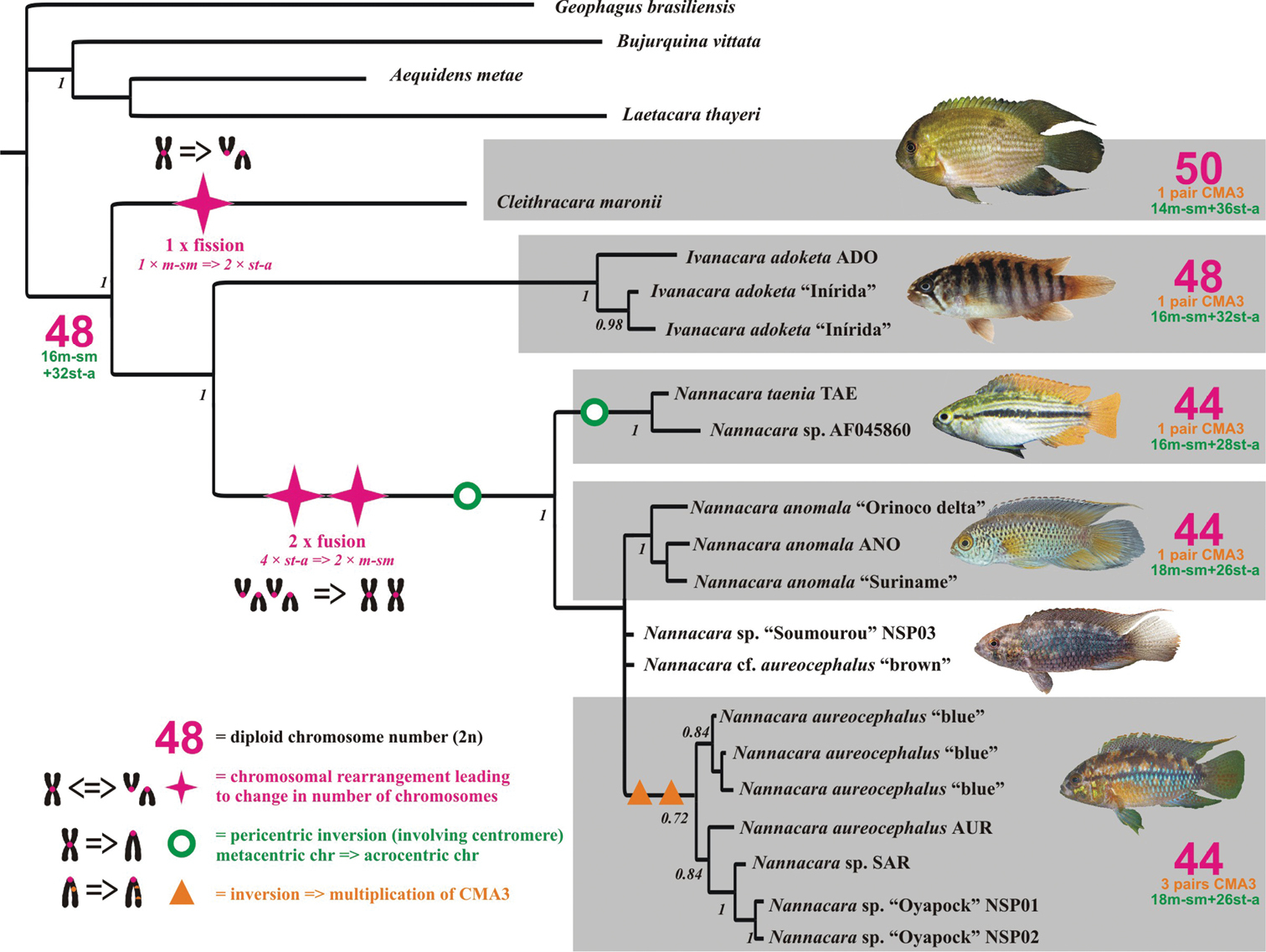
Phylogenetic reconstruction based on the DNA sequences of up to four genes shows monophyly of the genus Nannacara (three species used in this study) and its sister relationship with the genus Ivanacara (one species present in our study). The monotypic genus Cleithracara (Cleithracara maronii) represents then basal lineage to the rest of Nannacara + Ivanacara (Fig. 2). The observed karyotype characteristics, i.e. the diploid chromosome number, the karyotype and the phenotype, were mapped on the phylogenetic tree and allowed reconstruction of the scenario of genome/karyotype evolution in the studied cichlids as well as to reconstruct as well as of the most likely hypothetical karyotype of an ancestor of the whole group. An ancestral karyotype of 2n = 48 was hypothesized as (16m-sm + 32 st-a) and was estimated as a basal stage for the clade by the most parsimonious reconstruction based on our material. The ancestor also had most likely only one pair of CMA3 sites (Fig. 2).
Karyotype characteristics of the South American dwarf cichlids, including the diploid number of chromosomes (2n), chromosome categories, and CMA3 phenotype.
| Species | 2n | Karyotype | CMA3 signals |
|---|---|---|---|
| Cleithracara maronii | 50 | 14sm+36st-a | 1 sm pair |
| Ivanacara adoketa | 48 | 16m-sm+32st-a | 1 st-a pair |
| Nannacara anomala | 44 | 18m-sm+26st-a | 1 m-sm pair |
| Nannacara aureocephalus | 44 | 18m-sm+26st-a | 3 m-sm pair |
| Nannacara taenia | 44 | 16m-sm+28st-a | 1 st-a pair |
Karyotypes arranged from Giemsa stained chromosomes (left) of five species of cichlids: Cleithracara maronii, Ivanacara adoketa, Nannacara anomala, Nannacara aureocephalus, Nannacara taenia. Selected metaphases stained with Giemsa staining (center) and sequentially by CDD banding (right). White arrows indicate chromosomes with positive Chromomycin A3 signals. Bar=10µm.
Phylogenetic relationships of cichlid fishes of genera Nannacara, Ivanacara and Cleithracara. Phylogenetic tree reconstructed based on the mitochondrial (cytochrome b, 16S rRNA) and nuclear (S7, RAG1) genes. Karyotype characteristics, such as diploid chromosomal number (2n), karyotype formula and CMA3 phenotype were mapped on the tree and interpreted under the most parsimonious criterion. Ancestral karyotype of the group evolved from the ancestral cichlid karyotype 48st-a (
Two of the five species presented within this study have been previously studied in
In the clade of Neotropical cichlids, three trends in karyotype differentiation can be distinguished (
All of the species within the studied evolutionary lineage have a higher proportion of sub-metacentric chromosomes in their karyotypes compared with the rest of cichlids (
The CMA3 signals represent usually the GC-rich DNA segments of heterochromatic regions, often correlated with the location of active or inactive NORs, usually represented by the rDNA regions in genome (
After
The phylogenetic reconstruction of the Nannacara – Ivanacara – Cleithracara clade (also called NIC clade in
Within Nannacara aureocephalus, more distinct forms are known; some of them were introduced into the aquarium trade under different names. So far no robust revision of Nannacara is available, and it is therefore difficult to make any taxonomic conclusion based on our data set. However, at least two different forms of Nannacara aureocephalus are spread among the aquarium hobbyist within Central Europe (Germany, Poland, Czech Republic, Slovakia) – one of them called “blue” and the other one called “brown” both included in our analyses. These forms are not of artificial origin, as usually F1 progeny of the wild caught individuals has been studied. Intuitively, the blue morph shows more light-blue coloration with iridescent elements both on the face and body, while the “brown” form doesn’t have the iridescent coloration and possess darker brown to dark-green coloration pattern. We have shown that those two morphs are genetically distinct; however, more detailed future work is necessary on this species/genus.
Cichlid karyotypes show some general common features - for example many species from African and Neotropical cichlids possess one pair of significantly larger chromosomes. Although the homology of the largest chromosome within the African lineage has been proved (
Although all the studied species from the Nannacara – Ivanacara – Cleithracara clade are characterized by the karyotype “B” (
The proposed mechanisms of chromosomal rearrangements are described in cichlids as well as in other fish species. Usually the sub-metacentric chromosome arises during the (centric) fusion, when two acrocentric-telocentric chromosomes fuse (
To conclude, we aimed to provide a comparative study on a small scale of three genera combining molecular and cytogenetic approaches. Assuming that cytogenetic data provide additional information, which is undetectable by molecular genetics (
We would like to thank Jan Nekola, Wolfgang Staeck, Tomáš Kučera, Ingomar Kranz, Leonel Calderón for providing of the samples or live specimens. We would like to thank Martina Pokorná and Marie Rábová for their constructive comments on the preliminary results. We thank Carlos Ziok, Jaroslav Hofmann and Miloslav Petrtýl for providing us their photos of the fish. The project was supported by S grant of MŠMT ČR and CIGA 20132016.